Ion Channels and Regulators
About Ion Channels and Regulators
Ion channels are specialized protein channels located in the cell membrane of all living organisms. These channels allow the selective passage of ions, such as sodium, potassium, calcium, and chloride, across the membrane. The regulation of ion flow is essential for various physiological processes, including cellular communication, muscle contraction, and nerve impulse transmission.
Ion channels provide a means for ions to move across the cell membrane, which is typically impermeable to charged particles. These channels can be gated, meaning they can open and close in response to specific stimuli. The gating of ion channels allows for precise control of ion movement and helps maintain the delicate balance of ions inside and outside the cell.
Regulators of ion channels play a crucial role in modulating their activity. These regulators can be other proteins, signaling molecules, or even changes in membrane potential. They can influence the opening and closing of ion channels, affecting the flow of ions across the membrane. By regulating ion channels, these molecules and mechanisms can fine-tune cellular responses to various stimuli.
The importance of ion channels and regulators is evident in various physiological processes. For example, the action potential in neurons relies on the opening and closing of ion channels to propagate electrical signals along the axon. In muscle cells, ion channels control the contraction and relaxation of muscles by modulating the influx and efflux of calcium ions.
Understanding ion channels and their regulators is critical in the development of therapeutics and drugs. Dysregulation of ion channels can lead to various diseases, including channelopathies, which are characterized by abnormal ion channel function. By uncovering the mechanisms of these channels and regulators, researchers can identify potential drug targets and design interventions to maintain or restore normal cellular function.
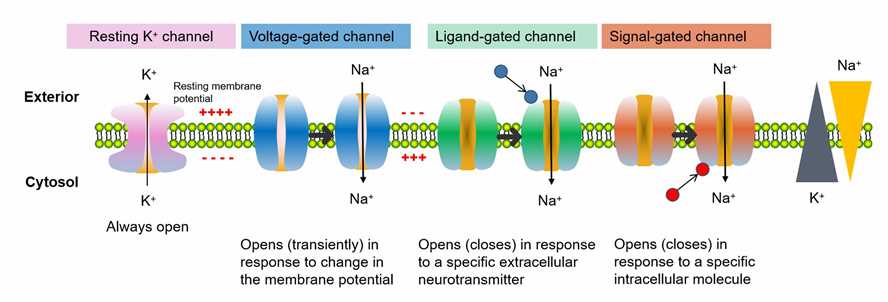
Biological Functions of Ion Channels and Regulators
Ion channels and regulators play crucial biological functions in various physiological processes. Here are some of their key functions:
Neuronal Signaling: Ion channels are essential for generating and propagating electrical signals in neurons. Voltage-gated ion channels, such as sodium (Na+), potassium (K+), and calcium (Ca2+) channels, allow the flow of ions across the neuronal membrane, enabling the rapid changes in membrane potential necessary for action potential generation and transmission. Ligand-gated ion channels, like glutamate and GABA receptors, mediate synaptic transmission and regulate neuronal excitability.
Muscle Contraction: Ion channels are vital for muscle contraction in both skeletal and cardiac muscles. Calcium ions play a central role in muscle contraction. Voltage-gated calcium channels in skeletal muscle cells are responsible for triggering calcium release from the sarcoplasmic reticulum, leading to muscle fiber contraction. In cardiac muscle, calcium influx through voltage-gated calcium channels initiates the contraction of the heart.
Hormone Secretion: Ion channels are involved in the secretion of hormones from endocrine glands. For example, voltage-gated calcium channels in pancreatic beta cells are crucial for insulin release in response to elevated blood glucose levels. Ion channels also contribute to the release of other hormones, such as adrenaline, from adrenal glands.
Fluid and Electrolyte Balance: Ion channels and regulators are essential for maintaining proper fluid and electrolyte balance in the body. Epithelial ion channels, including sodium, potassium, and chloride channels, regulate the transport of ions across epithelial tissues such as the kidneys, lungs, and intestines. These channels play a critical role in processes like reabsorption, secretion, and osmotic balance.
Cardiac Rhythm: Ion channels are responsible for generating and maintaining the rhythmic electrical activity of the heart. Specialized ion channels, including sodium, potassium, and calcium channels, regulate the action potential duration and shape in cardiac cells. Dysfunction or abnormalities in these channels can lead to arrhythmias and other cardiac disorders.
Sensory Perception: Ion channels are involved in sensory perception, allowing organisms to detect and respond to various stimuli. For example, ion channels in sensory neurons of the auditory system are responsible for transducing sound waves into electrical signals. Similarly, ion channels in photoreceptor cells of the retina mediate the conversion of light stimuli into visual signals.
Cell Volume Regulation: Ion channels and transporters play a role in maintaining cell volume and osmotic balance. Channels such as aquaporins facilitate the movement of water across cell membranes, while ion channels like potassium channels regulate ion transport to maintain proper cell volume and prevent cell swelling or shrinkage.
Immune Response: Ion channels and regulators are involved in immune cell function and the immune response. Potassium channels, for example, play a role in the activation and proliferation of immune cells. Calcium channels are also important for immune cell signaling and cytokine release.
Nutrient Uptake: Ion channels and regulators facilitate the uptake of essential nutrients, such as sodium, potassium, calcium, and magnesium, into cells. For example, sodium channels are involved in the absorption of sodium ions in the intestines and kidneys.
Hormone Secretion: Ion channels and regulators are involved in the release of hormones from endocrine cells. Calcium channels, in particular, play a critical role in the exocytosis of hormone-containing vesicles from endocrine cells.
Cell Proliferation and Differentiation: Ion channels and regulators are also implicated in cell proliferation and differentiation processes. For instance, specific potassium channels are involved in regulating cell proliferation and differentiation in various tissues and organs.
These are just some of the many biological functions of ion channels and regulators. Their diverse roles highlight their importance in numerous physiological processes, and dysregulation of ion channels can contribute to various diseases and disorders. Understanding the mechanisms and functions of ion channels is crucial for advancing our knowledge of biological processes and developing targeted therapies for related conditions.
The Significance of Studying The Ion Channels and Regulators
Studying ion channels and regulators is significant for several reasons:
1. Understanding Cellular Function: Ion channels play a crucial role in maintaining the electrical activity of cells. They control the movement of ions, such as calcium, sodium, and potassium, across cell membranes, which is essential for various cellular functions, including nerve transmission, muscle contraction, and hormone secretion. By studying ion channels, scientists can gain insights into how these processes are regulated, helping them understand normal cellular function and dysfunction in diseases.
2. Disease Research: Dysfunctional ion channels are responsible for a wide range of diseases, including neurological disorders (e.g., epilepsy, Parkinson's disease), cardiac arrhythmias, cystic fibrosis, and diabetes. Investigating ion channels and regulators provides vital information about the underlying mechanisms of these diseases. It helps identify potential therapeutic targets and develop drugs that modulate ion channel activity, leading to more effective treatments.
3. Pharmacology and Drug Development: Ion channels are targets for many drugs, including painkillers, antihypertensives, and antiarrhythmics. Studying ion channels allows researchers to understand the structure, function, and regulation of these targets. This knowledge is instrumental in developing new drugs that can modulate ion channel activity specifically, resulting in safer and more effective medications.
4. Biotechnology and Bioengineering: Ion channels have important applications in biotechnology and bioengineering. For example, they are used in drug discovery platforms to screen compound libraries or test drug candidates' effects on ion channel activity. Ion channels are also utilized in bioelectronic devices, such as biosensors or artificial tissues, where their specific properties and regulatory mechanisms are harnessed for various applications.
5. Evolutionary Studies: Ion channels and regulators have evolved throughout species, adapting to different environments and playing critical roles in physiological processes. By studying ion channels and regulators, scientists can gain insights into the evolution of these proteins and how they have contributed to the diversity of life forms on Earth. This knowledge can help shed light on the origins and development of complex biological systems.
Available Resources for Ion Channels and Regulators
With a commitment to providing high-quality products and services, Creative BioMart is a leading supplier in the life sciences research industry. Our extensive catalog includes a diverse range of recombinant proteins related to ion channels and regulators, offering researchers convenient access to essential tools for their studies. Our recombinant proteins related to ion channels and regulators are meticulously produced using advanced recombinant protein expression technologies. In addition to recombinant proteins, we also provide cell and tissue lysates from various species. These lysates offer a convenient source of proteins for various applications, including Western blotting, protein-protein interaction studies, and enzymatic assays. To facilitate protein purification and enrichment, we offer protein pre-coupled magnetic beads. These magnetic beads are functionalized with specific ligands to selectively capture and isolate target proteins from complex biological samples. This allows for efficient purification and enrichment of proteins of interest, saving researchers time and effort in their experiments.
The following are ion channel and regulator related molecules. Click on a molecule/target to view extensive resources.
At Creative BioMart, we understand that every research project is unique and may require customized solutions. Therefore, we offer customized services tailored to meet specific research needs. Our team of experienced scientists is dedicated to providing personalized support and guidance throughout the entire research process. Whether it's protein expression, purification, characterization, or assay development, we are committed to assisting researchers in achieving their goals.
If you have any questions or customization needs, please feel free to contact our customer service team. We look forward to working with you to jointly promote scientific research progress!


