AMPK (AMP-activated Protein Kinases)
Autophagy, also known as type II programmed cell death, is a lysosomal degradation pathway essential for cell survival. So far, many studies have suggested that autophagy is an antiapoptotic cell protection mechanism used by multiple cells. Autophagy is an evolutionarily conserved lysosome-dependent catabolic process This degradation process exerts a crucial influence on multiple physiological processes such as cellular homeostasis, aging, development, host-pathogen interactions, differentiation, and cell death and survival. In the last two decades, autophagy-related pathways and autophagy related genes (ATGs) have been extensively studied in yeast and in humans as model organisms.
Autophagy occurs under basal conditions in most eukaryotic cells, allowing the degradation and recycling of surplus and damaged cellular components. In diseases, dysfunction of the autophagy process has been linked to several pathological conditions. This connection stems from autophagy’s role in quality control of the proteome, organelles, and the cellular energetic balance. Autophagy is activated in response to cellular damage and more challenging insult-induced adaptive responses such as nutrient and growth factor withdrawal, infection, hypoxia, or heat can also trigger autophagy. This mechanism also plays a pivotal role during cellular starvation, producing an increase in ATP levels via protein breakdown. Some dying eukaryotic cells exhibit highly enhanced autophagic activity. This kind of cell lysis has been named Type 2 Cell Death (T2CD) due to its distinctive biological signature in relation to Type 1 Cell Death (T1CD) or apoptosis.
Classification of Autophagy
There are three types of autophagy—chaperone-mediated autophagy (CMA), macroautophagy (MA), and microautophagy (mA). The term “autophagy” usually indicates macroautophagy unless otherwise specified. The best characterized is MA, which in turn is frequently identified simply as “autophagy”. MA (hereafter referred to as autophagy, unless otherwise specified) can be divided into different phases: induction, elongation, maturation, transport to lysosomes, and degradation. A concise update on its mechanisms is shown on Figure. 1. mA is a form of cargo degradation in which targeted cytoplasmatic materials are directly engulfed by the vacuoles constituting multivesicular bodies (MVBs); subsequently, the cargo is cleaved by enzymes. CMA does not require the formation of MVBs; proteins are directly internalized in lysosomal lumen. This kind of autophagy is highly selective. It only captures proteins with the (Lys-Phe-Glu-Arg-Gln) KFERQ motif.
Process of Autophagy
The autophagy process can be divided into several steps: the induction of autophagy, the formation of autophagic bodies, the fusion of vesicles, and the decomposition of autophagic bodies. ATG protein has a key role in the formation and transport of autophagic bodies. Among the ATG protein, ATG1, ATG6, ATG8 (ULK1, Beclin1, and LC3 in mammals, respectively), and ATG5 are four major regulators of the autophagy pathway. ATG1 kinase domain has an important role in autophagy induction. In mammalian cells, the interaction of ATG13 with ULK1 and ULK2 can mediate the interaction of ULK protein with FIP200. ULK-ATG13-FIP200 complex is the direct target of mTOR and is an important regulator of mTOR signal transduction during autophagy (Figure 1). By inhibiting the activity of mTOR, the activity of ATG1 kinase is enhanced, and the surrounding related ATG proteins are inserted into the PAS (the site of autophagy formation) to promote the formation of ATG1-ATG13-ATG17 scaffold. The initial assembly of the bilayer membrane requires the III type phosphatidylinositol kinase (PtdIns3K) complex. After vesicle fusion, the degradation of autophagy depends on a series of lysosomal or vacuolar acid hydrolases, including protease A and B. In the case of cell starvation, the small molecules produced by amino acid degradation are transported back to the cytoplasm for protein synthesis and maintenance of cell function.
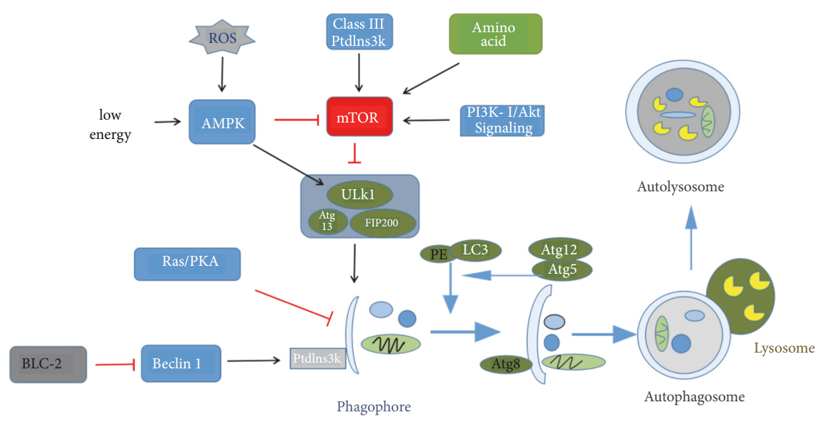 Figure 1. Process and regulatory mechanism of autophagy. (Pan Luo; et al. 2019)
Figure 1. Process and regulatory mechanism of autophagy. (Pan Luo; et al. 2019)
Regulatory Mechanism of Autophagy
mTOR is an atypical serine/threonine protein kinase involved in phosphoinositide 3-kinase- (PI3K-) related kinase family, which interacts with several proteins to form two different complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Previous studies have suggested that mTOR has a negative regulatory effect on autophagy. As mentioned above, mTOR can regulate signal transduction during autophagy through ULK-ATG13-FIP200 complex. Type III phosphatidylinositol enzyme (PI3K) and Beclin1 can be combined to form complexes for autophagosome formation, while BCL-2 and Beclin1 are combined with competitive inhibition of Beclin1 and PI3K to inhibit autophagy activity. Ras/PKA signaling pathway negatively regulates the autophagy activity by regulating the early steps of the Cvt transport process. Furthermore, recent studies have suggested that some nanoparticles may also affect the autophagy in cells.
References:
1. Pan Luo.; et al. The role of autophagy in chondrocyte metabolism and osteoarthritis: a comprehensive research review. BioMed Research International, 2019, 2019(4):1-7.
2. Lorenzo-Pouso.; et al. AI Autophagy in periodontal disease: Evidence from a literature review. Archives of Oral Biology, 2019,102;55-64.


