There are lots of great articles published on Cell in October, let’s share some highlights for you.
1. New discovery on cancer metabolic which challenge the view of nearly 100 years
Brandon Faubert, Kevin Y. Li, Ling Cai et al. Lactate Metabolism in Human Lung Tumors. Cell, 5 October 2017, 171(2):358–371, doi:10.1016/j.cell.2017.09.019
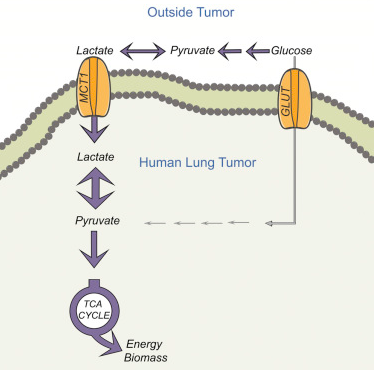
In this new study, researchers from the Children’s Medical Center (CRI) at the University of Texas Southwestern Medical Center (CRI) found that lactic acid supplies fuel to growing tumors. This finding challenged the Warburg effect which lasted for nearly a century.
The Warburg effect, named after the German cancer biologist Otto Warburg, has three main aspects: most cancer cells have (1) rapid glucose uptake; (2) lactic acid fermentation occurs even in the presence of oxygen; (3) lactose secrets as one kind of the waste.
In this study, these researchers confirmed that lactic acid was not only a waste but also as a fuel that was ingested by lung cancer cells grown in patients and mice. According to a study published previously in the Cell (Cell, 11 February 2016, doi: 10.1016 / j.cell.2015.12.034), DeBerardinis Laboratories has demonstrated that glucose oxidation is activated in tumors. Combined with this finding, the results of this new study pose a challenge to the Waller effect theory.
“We believe that lactic acid is one of the fuels that promote lung cancer cell growth, proliferation, and possibly even metastasis,” DeBerardinis said. “Cancer metabolism is clinically maneuverable and understanding lactate pathways may help us to identify the targets for lung cancer treatment. When using the imaging tracer, lactic acid uptake may also be predictive.”
2. New hope for Diabetes! New insulin sensitizers identified
Fanny Langlet, Rebecca A. Haeusler, Daniel Lindén et al. Selective Inhibition of FOXO1 Activator/Repressor Balance Modulates Hepatic Glucose Handling. Cell, Published online: October 19, 2017, doi:10.1016/j.cell.2017.09.045
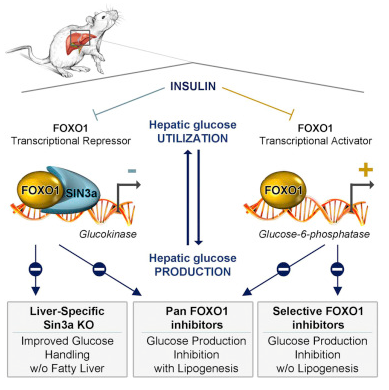
Type II diabetes originated from the insulin resistance (insulin resistance), that is, cells stop react with this hormone. Restoring insulin sensitivity is an effective strategy for the prevention and treatment of this diabetes. But the drug on the market that replicates insulin-sensitive also promotes lipid (fat) production, which can lead to some serious side effects.
In a new study, researchers from Columbia University, Mount Sinai School of Medicine and AstraZeneca, Sweden, found that it is available to improve insulin sensitivity while avoiding these side effects. Relevant findings were published online October 19, 2017, in the Journal Cell, titled “Selective Inhibition of FOXO1 Activator / Repressor Balance Modulates Hepatic Glucose Handling.”
One method that has been studied before is to inhibit the protein FOXO1. Animal studies have shown that the liver produces less glucose when FOXO1 is inhibited. But like other insulin sensitizers, inhibiting FOXO1 also promotes lipid production. “Therefore, treating insulin resistance with a broad-based FOXO1 inhibitor can result in a range of unwanted side effects such as weight gain. Unfortunately, for FOXO1 insulin sensitivity drug, you have to bear both good and bad.”
In the current study, the researchers found a method to partially suppress FOXO1, resulting in glucose levels reduction but no change in lipid levels. “We need to understand how these two FOXO1 regulatory mechanisms discriminate with each other, so we can identify selective inhibitors,” said Daniel Lindén, co-author of the paper and a scientist with the biotechnology department for innovative drugs and early development at AstraZeneca.
In a study conducted in mice, Dr. Accili and his colleagues found that FOXO1, along with the protein SIN3A, limits lipid production. “This suggests that if we can identify one molecule that acts on FOXO1 during the glucose-producing process without interfering SIN3A, we can improve insulin sensitivity and lower blood glucose without increasing fat levels,” Dr. Accili said.
3. Open the genetic loop in cancer cells and trigger an immune attack
Lior Nissim, Ming-Ru Wu, Erez Pery et al. Synthetic RNA-Based Immunomodulatory Gene Circuits for Cancer Immunotherapy. Cell, Published online: October 19, 2017, doi:10.1016/j.cell.2017.09.049
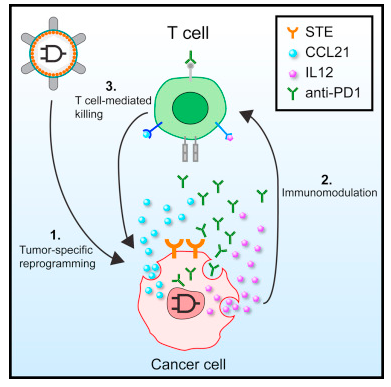
In a new study, researchers from the Massachusetts Institute of Technology (MIT) developed a synthetic gene circuit that activates the body’s immune system to attack target when it detects signs of cancer. This gene circuit activates a therapeutic response only when it detects two specific cancer markers. Relevant findings were published online in the Cell journal on October 19, 2017.
Lu and his team, including MIT postdoctoral fellows Lior Nissim and Ming-Ru Wu, built a genetic loop that codes for DNA in cancer cells and non-cancer cells. This gene circuit can be customized to respond to different tumors. It is based on the simple AND gates used in electronics. This AND gate opens only when both inputs exist.
The difference between cancer cells and normal cells lies in their gene expression profile. Thus, these researchers developed synthetic promoters that code for this gene loop-DNA sequences that initiate gene expression in cancer cells only. By virus, this gene loop is transported to the affected area in the body. Certain proteins that are active in tumor cells then bind to these synthetic promoters and activate them. This gene loop is only turned on when both of these cancer promoters are activated. This allows the gene circuit to target the tumor more accurately than existing therapies because it requires the presence of both cancer-specific signals before it can react.
When the researchers tested the gene circuit in vitro, they found it was able to identify ovarian cancer cells from other non-cancerous ovarian and other cell types. They then tested the gene circuit in mice that underwent ovarian cancer cell transplantation and confirmed that it triggers the T cells to find and kill these cancerous cells without damaging the other cells around them.
4. Gut bacteria from wild-type mice improve the health of laboratory mice
Stephan P. Rosshart, Brian G. Vassallo, Davide Angeletti et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell, Published online:October 19, 2017, doi:10.1016/j.cell.2017.09.016
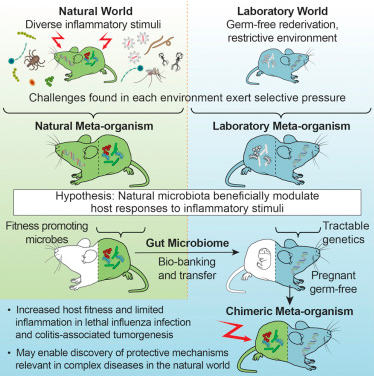
In a new study, researchers reported that laboratory mice that received gut bacterial translocation in wild mice were significantly high survival rate when infected deadly influenza virus and confronted colorectal cancer than lab mice that had their own gut bacteria. Relevant findings were published online in the Cell journal on October 19, 2017, and the paper was titled “Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance.” The paper was co-authored by Barbara Rehermann, Ph.D., director of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Liver Diseases Laboratory and Stephan Rosshart, Ph.D., an NIDDK postdoctoral fellow.
Laboratory mice were carefully nurtured, fed and housed under carefully controlled conditions, resulting in predictable traits and genetic characteristics for each laboratory mouse. This is a big advantage in basic biology, but producing this predictability means the controlled environment, instead of the pressure of survival from the outside world, affects the microbiota of laboratory mice. So the researchers tried to recover what they lost: gut microbiota in naturally co-evolving wild mice. They captured more than 800 wild mice in eight places in Maryland and the District of Columbia, from which to find healthy, fit candidates to provide their gut bacteria.
They then tested and compared the gut microbiome of the wild mouse (Mus musculus domesticus) and common laboratory mouse strain C57BL/6 obtained from various sources. They confirmed that C57BL/6 mice had a significantly different gut microbiome from wild-type mice.
These researchers subsequently transplanted intestinal flora of wild mice into pregnant, sterile C57BL/6 mice. These sterile mice grew up in a sterile environment without their own microbiome. For comparison, they also transplanted gut flora from a normal-cultured C57BL /6 mouse into a group of pregnant, sterile C57BL/6 mice as a control group. After four generations these mice still either carried the gut microbiome of wild mice or carried the gut microbiome of laboratory mice inherited from their maternal ancestry.
When exposed to high doses of influenza virus, 92% of laboratory mice carrying the gut microbiome of wild mice survived, however, only 17% of laboratory mice survived in the control group. In other experiments, laboratory mice harboring the gut microbiome of wild-type mice have better outcomes in the treatment of induced colorectal tumors, however, as the control group, the experimental mice have more serious diseases. The beneficial effects of gut flora in wild mice are associated with reduced inflammation in both mouse models.
(to be continued…)