Protein-Lipids/Nucleic Acid Interaction Assay and Screening
Protein interactions with lipids and nucleic acids play essential roles in cellular regulation, signaling pathways, gene expression, and disease mechanisms. At Creative BioMart, we provide comprehensive protein-lipids and protein-nucleic acid interaction assay and screening services, enabling precise identification, mapping, and characterization of these critical molecular events. Leveraging advanced technologies such as ChIP, CLIP, EMSA, and Fat Western, our platform supports both fundamental research and translational applications. From transcription factor binding to RNA regulatory networks and lipid-binding specificity, we deliver high-quality, reproducible data tailored to your research goals.
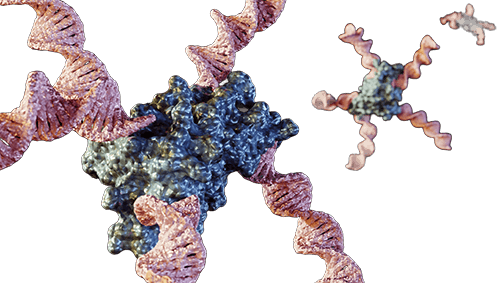
Background: Understanding Protein-Lipid and Protein-Nucleic Acid Interactions
Proteins rarely function in isolation. Their interactions with DNA, RNA, and lipids form the foundation of diverse biological processes, from transcriptional control and post-transcriptional regulation to membrane signaling and cellular metabolism. Dysregulation of these interactions is often implicated in cancers, neurological disorders, autoimmune diseases, and metabolic syndromes. Therefore, precise assays to study protein-lipid and protein-nucleic acid interactions are critical for understanding molecular mechanisms and identifying potential therapeutic targets.

Figure 1. A: An example of lipid-protein interaction. (Molina et al., 2015) B: An example of protein–DNA interaction. (Wang et al., 2013).
As a trusted partner in biotechnology services, Creative BioMart provides a complete suite of assays to study these interactions with accuracy, flexibility, and scientific rigor.
Protein-Lipid and Protein-Nucleic Acid Interaction Assay Services
Our Service Offerings
|
Chromatin Immunoprecipitation (ChIP) Investigates protein-DNA interactions in vivo, determining whether proteins (e.g., transcription factors) are bound to specific genomic regions. Also maps histone modifications to reveal chromatin regulation. |
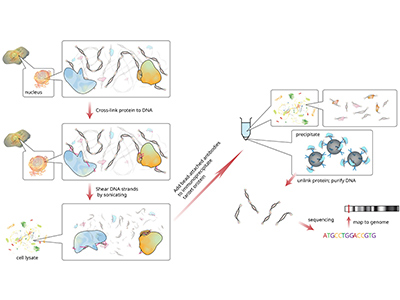 |
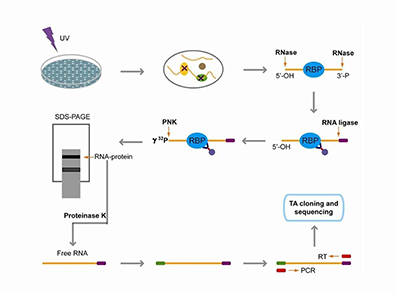 |
Cross-linking Immunoprecipitation (CLIP) Analyzes protein-RNA interactions by combining UV cross-linking with immunoprecipitation. Enables mapping of RNA-binding sites genome-wide, advancing understanding of post-transcriptional regulatory networks. |
|
Electrophoretic Mobility Shift Assay (EMSA) Detects protein-DNA or protein-RNA complexes, identifies binding specificity, and distinguishes between single- and multi-protein binding events. |
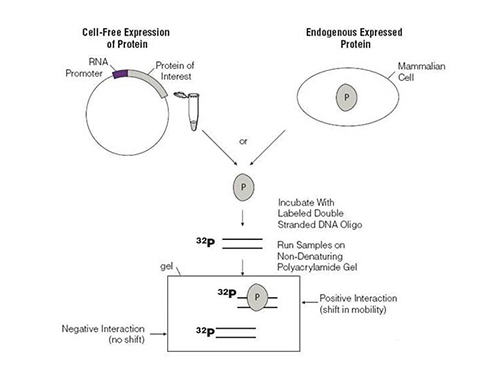 |
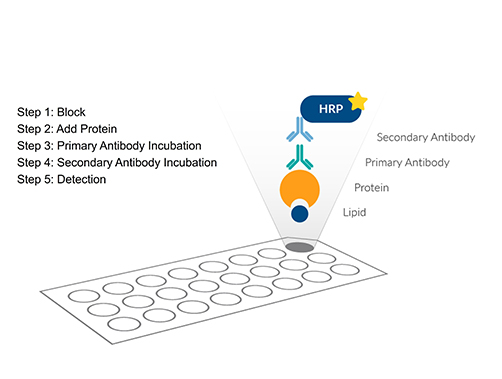 |
Fat Western Screens protein-lipid interactions by dot-blotting selected lipids. Identifies lipid-binding specificity and domains, estimates affinity constants, and supports signaling studies. |
|
Short Tandem Repeat (STR) Profiling Provides sample authentication and ensures experimental integrity. |
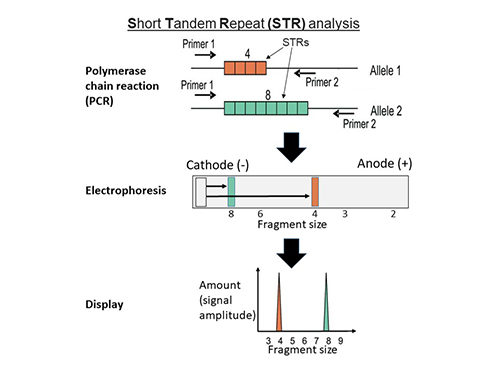 |
Service Workflow

Advantages of Choosing Creative BioMart for Interaction Analysis
- Comprehensive Portfolio: Wide range of assays covering protein-DNA, protein-RNA, and protein-lipid interactions.
- Advanced Techniques: State-of-the-art methodologies such as CLIP and Fat Western for accurate and sensitive results.
- Customization: Flexible solutions tailored to diverse project requirements and research objectives.
- Quality Assurance: Rigorous STR profiling and QC measures ensure experimental reliability.
- Collaborative Support: Expert consultation, experimental design assistance, and training are available.
- Proven Expertise: Trusted by academic and industry partners worldwide, with results supporting publications and therapeutic programs.
Case Studies in Protein-Lipid and Protein-DNA/RNA Screening
Case 1: Concurrent RNA-binding protein interactions revealed
Klass et al. , 2013. doi:10.1101/gr.153031.112
Understanding how RNA-binding proteins (RBPs) shape mRNA fate requires mapping not just their individual targets but also their concurrent interactions. The researchers developed a quantitative mass spectrometry–based method, combined with RNase treatment of affinity-purified RNA-protein complexes, to identify proteins binding simultaneously with an RBP of interest. Applied to Pab1, Nab2, and Puf3 in yeast, the method enriched for known RBPs, uncovered over 100 new candidates, and confirmed direct RNA binding in 77% of tested proteins. Results revealed novel RNA-binding roles, clarified shared target interactions, and distinguished concurrent binding from indirect associations, advancing insights into post-transcriptional regulation.

Figure 2. SDS-PAGE analysis of protein fragments resulting from partial digestion of Mbf1 with the protease chymotrypsin. The lanes from left to right contain the ladder, chymotrypsin only, the supernatant of protein fragments liberated by chymotrypsin digestion of Mbf1, and the protein fragments remaining on the beads. (Klass et al., 2013)
Case 2: Lipid binding specificity governs RAP1 membrane organization
Araya et al., 2024. doi:10.1021/jacs.4c02183
RAP1 proteins, members of the RAS small GTPase family, act as molecular switches by cycling between GDP- and GTP-bound states. Using high-resolution imaging, the researchers discovered that RAP1A and RAP1B form distinct nanoclusters on the plasma membrane, with further segregation depending on their activation state. Their C-terminal polybasic domains (PBDs) exhibit unique lipid binding specificities, which can be altered by single-point mutations. Molecular dynamics simulations confirmed that such mutations weaken interactions with phosphatidylserine, disrupting membrane association. These findings demonstrate that PBD-encoded lipid binding specificity dictates nanoclustering of RAP1 proteins, revealing a broader principle of small GTPase membrane organization.

Figure 3. Differential lipid sorting specificities are encoded in the membrane anchors of RAP1 proteins. (A,B) PM sheets of BHK cells co-expressing RFP-tagged wild-type (RFP-RAP1-WT) or GTP-bound mutant RAP1 (RFP-RAP1-G12V) with a GFP-tagged lipid probe for PS (GFP-LactC2), PIP2 (GFP-PLCδ), PIP3 (GFP-AKT), PA (GFP-PASS), or cholesterol (GFP-D4H) were labeled with 6 nm gold-anti-GFP and 2 nm gold-anti-RFP and imaged by EM. (Araya et al., 2024)
What Our Clients Say
“We collaborated with Creative BioMart to investigate transcription factor binding in colorectal cancer models using ChIP assays. Their team delivered highly reproducible mapping of promoter regions and histone modifications, which provided key insights into our epigenetic drug program. The data quality was exceptional and passed all of our internal validation steps. Their ability to handle complex chromatin samples really set them apart.”
— Director of Oncology Research | Global Pharmaceutical Company
“Our lab needed to identify RNA-binding sites of a splicing regulator implicated in neurodegenerative disease. Creative BioMart applied their CLIP expertise to generate precise, genome-wide binding profiles. Their combination of technical skill and responsive communication was impressive.”
— Principal Investigator | Leading University Medical School
“As a startup focused on metabolic disorders, we relied on Creative BioMart to screen lipid-binding domains of our lead signaling protein using Fat Western assays. They identified a strong affinity for phosphatidylserine, which reshaped our therapeutic strategy and helped secure Series A funding. Their quick turnaround and detailed reporting were invaluable in a fast-moving project environment.”
— Chief Scientific Officer | Biotech Startup
“We approached Creative BioMart with a complex project involving both EMSA and STR profiling to authenticate samples while studying transcription factor-DNA interactions. They customized the workflow to meet our requirements and ensured the data was robust enough for regulatory submission. Their attention to detail and willingness to adapt the assays to our needs saved us considerable time and resources.”
— R&D Director | Mid-Sized Pharmaceutical Company
FAQs About Protein-Lipid and Protein-Nucleic Acid Interaction Assays
-
Q: What types of interactions can your assays detect?
A: We provide comprehensive solutions to study protein interactions with DNA, RNA, and lipids. Our assays include ChIP for protein-DNA binding, CLIP for protein-RNA binding, EMSA for nucleic acid-binding dynamics, and Fat Western for protein-lipid interactions. Together, these platforms allow us to characterize binding specificity, map binding sites, and estimate affinity constants. -
Q: Can you work with complex or challenging samples?
A: Yes. Our team has extensive experience handling challenging biological materials, including chromatin from tissue samples, RNA-protein complexes with low abundance, and proteins with multiple lipid-binding domains. We optimize protocols for each project to ensure maximum sensitivity and reproducibility. -
Q: How do you ensure data reliability and reproducibility?
A: We use well-established methodologies, rigorous controls, and advanced detection systems such as HPLC, UV cross-linking, and immunoblotting to ensure consistent results. Each assay undergoes internal validation before reporting, making our data suitable for both research publications and regulatory submissions. -
Q: Do you provide customized workflows?
A: Absolutely. Whether you need genome-wide mapping of transcription factor binding, identification of lipid-binding specificity, or combined assay approaches, we tailor workflows to match your project goals. We also integrate services like STR profiling for sample authentication when required. -
Q: What kind of support do you provide beyond the assay itself?
A: In addition to performing the assays, we assist with experimental design, provide training upon request, and deliver comprehensive data interpretation.
Resources
Related Services
References:
- Araya MK, Chen W, Ke Y, et al. Differential lipid binding specificities of RAP1A and RAP1B are encoded by the amino acid sequence of the membrane anchors. J Am Chem Soc. 2024;146(29):19782-19791. doi:10.1021/jacs.4c02183
- Klass DM, Scheibe M, Butter F, Hogan GJ, Mann M, Brown PO. Quantitative proteomic analysis reveals concurrent RNA–protein interactions and identifies new RNA-binding proteins in Saccharomyces cerevisiae. Genome Res. 2013;23(6):1028-1038. doi:10.1101/gr.153031.112
- Molina ML, Giudici AM, Poveda JA, et al. Competing lipid-protein and protein-protein interactions determine clustering and gating patterns in the potassium channel from Streptomyces lividans (KCSA). Journal of Biological Chemistry. 2015;290(42):25745-25755. doi:10.1074/jbc.M115.669598
- Wang T, Sun HL, Cheng F, Zhang XE, Bi L, Jiang T. Recognition and processing of double-stranded DNA by ExoX, a distributive 3′–5′ exonuclease. Nucleic Acids Research. 2013;41(15):7556-7565. doi:10.1093/nar/gkt495
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

