Protein Interaction (3) Yeast-Two-Hybrid Assay
Pioneered by Stanley Fields and Ok-Kyu Song in 1989, the yeast-two-hybrid (Y2H) technique was originally designed to detect protein–protein interactions using the GAL4 transcriptional activator of the yeast Saccharomyces cerevisiae. Nowadays, the Y2H screen remains one of the leading molecular tools to study protein-protein interactions in native intracellular conditions. This screen is not only cheap and easy to operate, but can also be carried out in almost any laboratory with results seen within a matter of days. In addition, compared to other methods used to study protein-protein interactions such as crosslinking and coimmunoprecipitation, the Y2H screen allows detecting weak and transient interactions that may not be normally detected in the former approaches, since the two hybrid system is a sensitive, in vivo method. Moreover, purified proteins or antibodies, or cell disruption are not required to Y2H. Most importantly, this approach makes it possible to screen gene libraries to identify and clone genes of previously unknown proteins that interact with a target protein.
This innovative technology employs the GAL4 transcriptional activator and takes advantage of the fact that GAL4 can be physically separated into two domains whose function can be re-established once the two domains are brought into proximity. Many eukaryotic transcription factors including GAL4 and LEXA have at least two distinct functional domains; one direct binding to specific DNA sequences (DNA binding domain or BD) and one activates transcription (activation domain or AD). In the classic Y2H screen two fusion proteins are required: a protein of interest fused to the DNA binding domain (BD) of GAL4 (also referred to as the bait protein) and the library protein(s) fused to the activation domain (AD) (also referred to as the prey protein). Interaction between the bait and prey protein brings the BD and AD of GAL4 close enough together to allow transcription of the downstream reporter genes that subsequently allows growth on selective media.
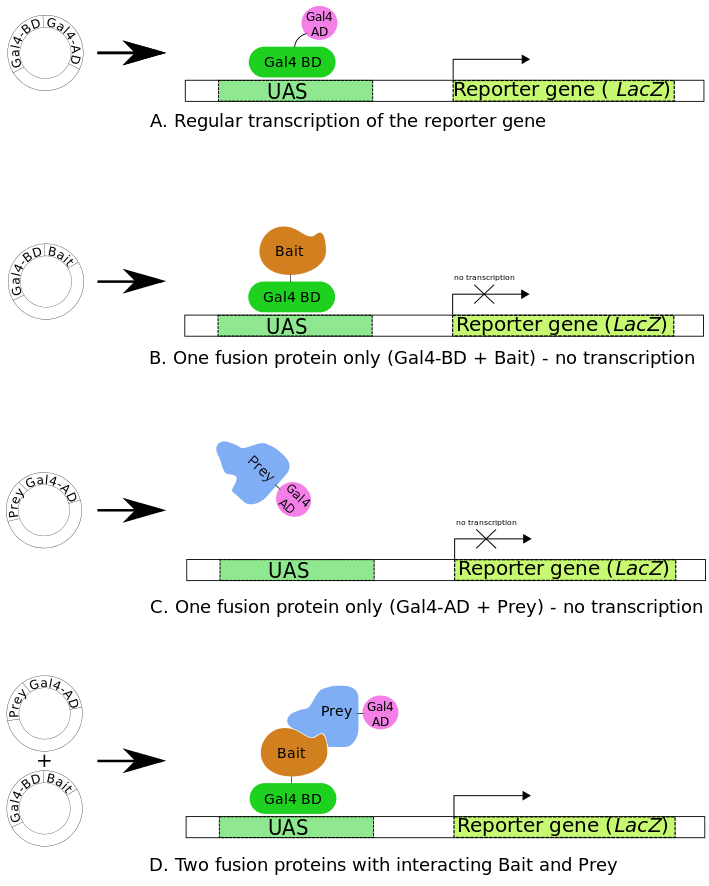
Fig1. Principle of Yeast-Two-Hybrid
However, although the classic Y2H screen is an attractive method to study a protein there are also some limitations. One limitation is that although this screen occurs within yeast cells and thus occurs under native cellular conditions, the bait and prey protein must both be able to translocate to the nucleus in order to allow transcription of the reporter gene. Therefore, interactions between the bait protein and membrane-bound, integral membrane and/or proteins localized to subcellular compartments cannot be detected and would generate what is called a false negative. Further, the bait and prey protein should not be able to autoactivate the reporter gene which would lead to the appearance of false-positives. False positives are one of the biggest drawbacks of the Y2H screen because the appearance of false positives may occur for multiple reasons, such as the one just mentioned or non-specific interactions. However, these limitations can be overcome. If the bait or prey protein autoactivates the reporter gene(s) then by using increasing concentrations of 3-amino-l,2,4-triazole (3AT) the expression of the HIS3 gene product (which is one of the reporter genes in the Y2H screen) is inhibited accordingly. To distinguish false positives from the real interactions, the identified interaction should be verified through other methods such as pull-down assays and co-immunoprecipitations. Finally, in order to overcome the generation of false negatives other Y2H systems have been developed: the SOS- and the RAS recruitment systems (SRS and RRS) (Hubsman et al., 2001) which detects interactions with membrane-bound proteins and the Split-ubiquitin system (Stagljar et al., 1998) which detects interactions occurring between cytosolic proteins and membrane proteins.
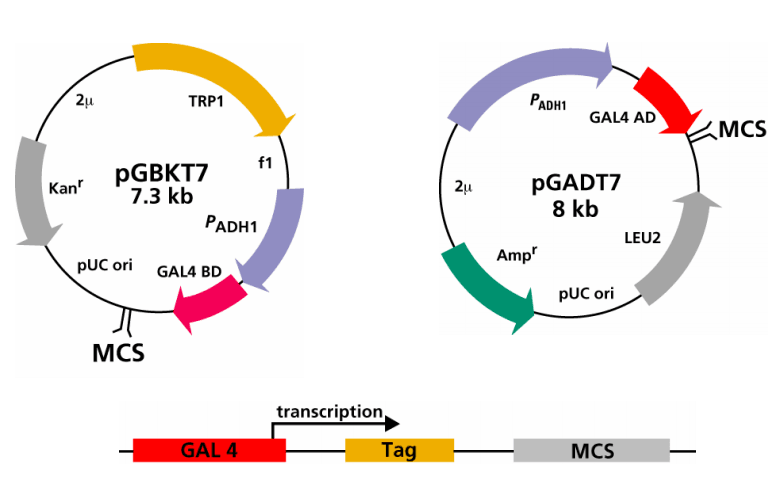 Fig2. Plasmids used in Y2H
Fig2. Plasmids used in Y2H
In other cases, yeast-two-hybrid system has been used to screen cDNA library against a protein of interest, thus identifying candidate genes encoding proteins which bind to the target protein. Only 1/6 of the library clones will be fused to the activation domain protein in the proper reading frame leading to biologically relevant clones, while 5/6 of the library clones will be out-of-frame. However, these latter clones represent a large collection of essentially random peptide fusions, which could be used to identify specific peptides with strong binding affinities to target protein.
For all the benefits of Y2H screening, this method has varieties of applications:
• Find new protein and new function of the protein;
• Determination of sequences crucial for interaction;
• Study the interaction between antigen and antibody in vivo;
• Screening the functional site of new drugs and the effection of the drug to the interaction of target protein;
• Establish genomic protein linkage map.
Reagents and Materials Required
• YPD or the appropriate SD liquid medium
• Sterile 1XTE/1X LiAc (Prepare immediately prior to use from 10X stocks)
• Sterile 1.5-ml microcentrifuge tubes for the transformation
• Appropriate SD agar plates (100-mm diameter)
• Appropriate plasmid DNA in solution
• Appropriate yeast reporter strain for making competent cells
• Carrier DNA
• Sterile PEG/LiAc solution (Prepare only the volume needed, immediately prior to use, from 10X stocks)
• 100% DMSO
• Sterile 1XTE buffer (Prepare from 10XTE buffer)
• Appropriate SD agar plates containing X-gal (80 mg/L) and 1X BU salts (Appendix C.A).
Yeast Transformation Procedures
- Inoculate 1 ml of YPD with several colonies, 2–3 mm in diameter.
- Vortex vigorously for 5 min to disperse any clumps.
- Transfer this into a ?ask containing 50 ml of YPD.
- Incubate at 30°C for 16–18 hr with shaking at 230 rpm to stationary phase (OD600>1.5).
- Transfer 30 ml of overnight culture to a ?ask containing 300 ml of YPD. Bring the OD600 up to 0.2–0.3.
- Incubate at 30°C for 3 hr with shaking at 230 rpm. At end point, the OD600 should be 0.4–0.6.
- Harvest cells in 50-ml tubes and centrifuge at 1,000 x g for 5 min at room temperature.
- Discard the supernatants and thoroughly resuspend the cell pellets in sterile TE or distilled H2O. Gather the cells into one tube.
- Centrifuge at 1,000 x g for 5 min at room temperature.
- Discard the supernatant.
- Resuspend the cell pellet in 1.5 ml of freshly prepared, sterile 1X TE/1X LiAc.
- Add 0.1 µg of plasmid DNA and 0.1 mg of carrier DNA to a fresh 1.5-ml tube and mix.
- Add 0.1 ml of yeast competent cells to each tube and mix well by vortexing.
- Add 0.6 ml of sterile PEG/LiAc solution to each tube and vortex at high speed for 10 sec to mix.
- Incubate at 30°C for 30 min with shaking at 200 rpm.
- Add 70 µl of DMSO. Mix well by gentle inversion.
- Heat shock for 15 min in a 42°C water bath.
- Chill cells on ice for 2 min.
- Centrifuge cells for 5 sec at 14,000 rpm at room temperature. Discard the supernatant.
- Resuspend cells in 0.5 ml of sterile 1X TE buffer.
- Plate 100 µl on each SD/–Leu/–Trp agar plate that will select for the desired transformants.
- Incubate plates, up-side-down, at 30°C until colonies appear (generally, 2–4 days).
- Pick several colonies from SD/–Leu/–Trp agar plate, streak on SD/–Leu/–Trp/-His agar plates and SD/–Leu/–Trp/–His /X-a-Gal agar plates separately.
- Incubate plates, up-side-down, at 30°C until colonies appear (generally, 2–4 days).
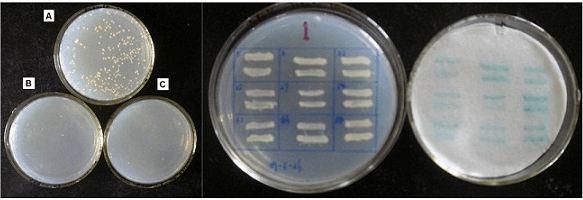 Fig3. Y2H assay results. Result on SD/–Leu/–Trp agar plates; result on SD/–Leu/–Trp/-His agar plates; result on SD/–Leu/–Trp/–His /X-a-Gal agar plates.
Fig3. Y2H assay results. Result on SD/–Leu/–Trp agar plates; result on SD/–Leu/–Trp/-His agar plates; result on SD/–Leu/–Trp/–His /X-a-Gal agar plates.
Creative BioMart provides yeast-two-hybrid screening, membrane-based yeast-two-hybrid system and mammalian two-hybrid system to analysis protein-protein interactions. We will be glad to discuss details of intended interaction studies with you and develop experimental strategies/methods tailored to your requirement. Please get in contact with Creative BioMart for more information or a detailed discussion.

