Uncategorized Sunday, 2025/11/23
This study has developed a universal and computationally guided ultra-precise base editing method, providing a promising platform for the mechanistic study and therapeutic correction of single-base mutations. Mitochondria are crucial for cellular energy production and overall cell function. Mutations in mitochondrial DNA (mtDNA) can cause debilitating diseases, particularly affecting high-energy-consuming tissues such as muscle and nerve cells. High-precision manipulation of mtDNA holds the promise of revealing underlying disease mechanisms and providing targeted therapeutic solutions, but achieving this level of precision remains a significant challenge.
In recent years, methods for editing mitochondrial DNA have emerged, but these methods often fail to achieve precise single-base editing and typically produce bystander editing, which has become a major limitation for the broader application of mitochondrial DNA editing technologies.
On November, a collaborative effort between the team of Peilong Lu from the School of Life Sciences at Westlake University and the team of Yangming Wang from the College of Future Technology at Peking University / Beijing RNA Research Center published a research paper titled "Computational design of a high-precision mitochondrial DNA cytosine base editor" in the Nature journal Nature Structural & Molecular Biology.
Based on an AI-assisted de novo protein design strategy, the study successfully developed a high-precision mitochondrial DNA cytosine base editor called DdCBE–TOD, which achieves single-base precision editing. This breakthrough paves a new path for the research and treatment of mitochondrial genetic diseases and provides an important strategy for the development of future high-precision gene editing tools.
Existing mitochondrial base editors typically produce bystander editing, a limitation that restricts the precision and therapeutic potential of mitochondrial DNA editing.
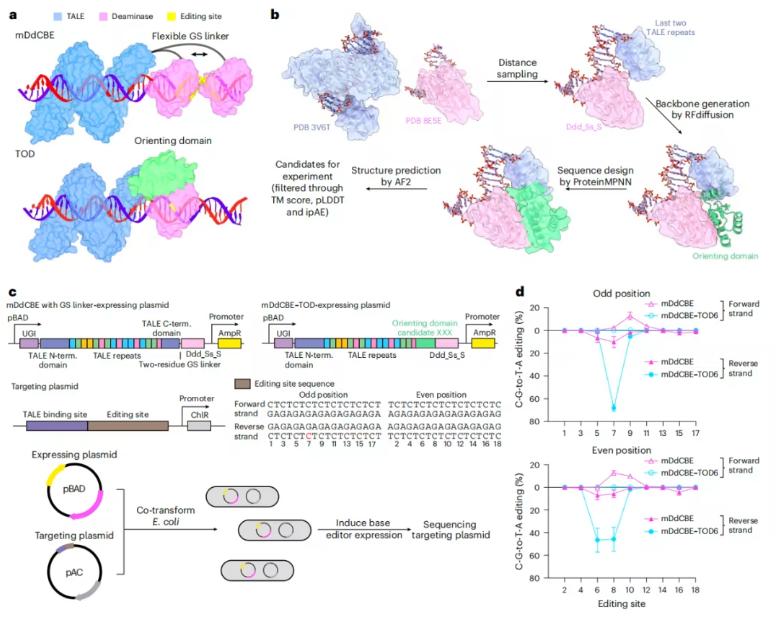
In this latest study, the research team proposed an AI-assisted de novo protein design strategy that creates a structurally rigid interface between the DNA-binding transcription activator-like effector (TALE) domain and cytosine deaminase, forming a unified editing module called TALE-directed deaminase (or TOD for short).
Cryo-electron microscopy structural analysis confirmed that the precise spatial structure of the TOD-DNA complex strictly confines the active window of the deaminase, thereby minimizing unnecessary deamination.
De Novo Design of Targeting Domains for Precise and Efficient Base Editing
To further enhance editing specificity, the research team developed a DddA-driven cytosine base editor-TOD (referred to as DdCBE–TOD), which almost completely eliminates off-target editing.
As a proof of concept, the research team applied DdCBE-TOD to generate mouse models of mitochondrial diseases and corrected pathogenic mtDNA mutations associated with MERRF syndrome in patient-derived cells, achieving single-base precision.
Overall, this study has developed a universal and computationally guided ultra-precise base editing method, providing a promising platform for the mechanistic study and therapeutic correction of single-base mutations.
Related Products & Services
- Protein Engineering Services
- Protein Interaction Service
- Protein Expression and Purification Services
- Drug Discovery Screening
- Protein Pathway Profiling
- Immune Checkpoint Proteins
- ADC Target Protein
- PROTAC Targets
- Cell and Gene Therapy
- Targets of CAR-T Cell Therapy
- Cancer Drug Targets
Reference
- Mi, L., Li, Y., Lv, X., Wan, Z., Liu, X., Zhang, K., Li, H., Yao, Y., Zhang, L., Xu, Z., Zhuang, X., Ji, K., Jiang, M., Wang, Y., & Lu, P. (2025). Computational design of a high-precision mitochondrial DNA cytosine base editor. Nature Structural & Molecular Biology, 1-12. https://doi.org/10.1038/s41594-025-01714-2
