Uncategorized Sunday, 2025/09/07
Groundbreaking Breakthrough! A Single Injection Alters Brain DNA, Offering Hope to Mice with Rare Neurological Diseases
DOI: 10.1016/j.cell.2025.06.038
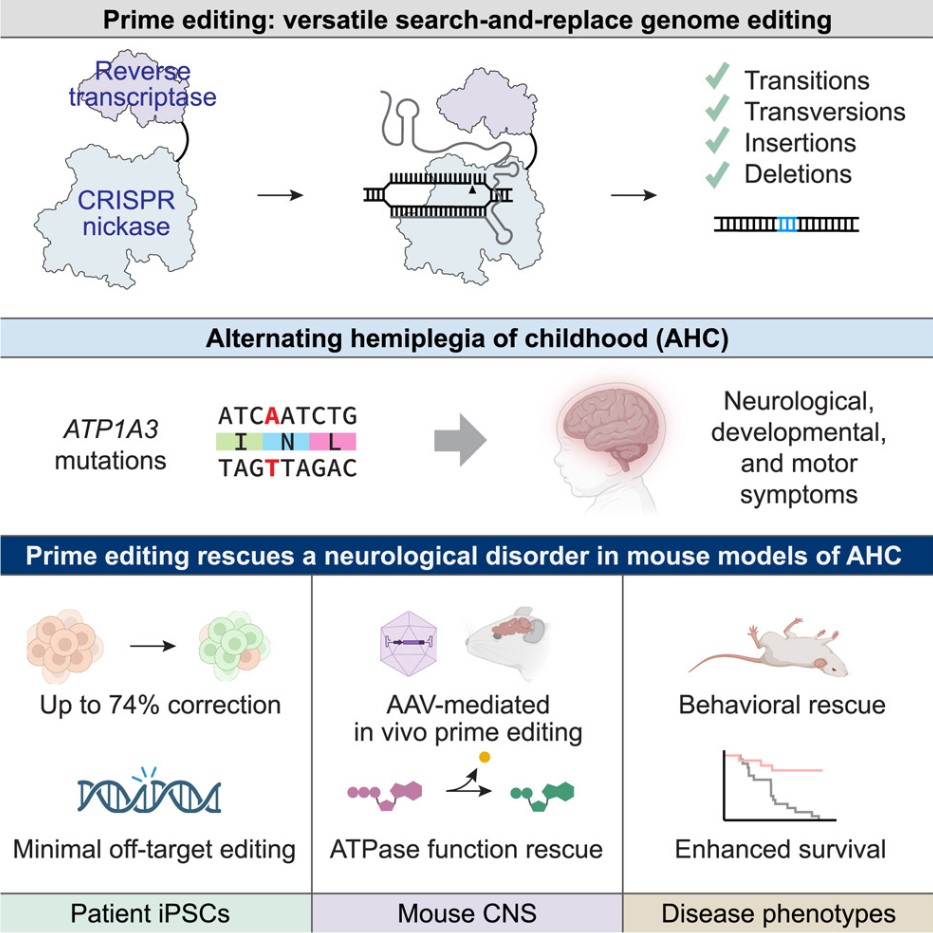
In the field of gene therapy, a result that could shake the academic community and patients alike has recently emerged. A team of elite researchers from the Jackson Laboratory (JAX), the Broad Institute, and the nonprofit RARE Hope collaborated to achieve a landmark medical exploration—directly editing DNA in the brains of mice through a single injection, successfully correcting the genetic mutation that leads to an ultra-rare disease. The related research results have been published in the top academic journal Cell, lighting up a beacon of hope for countless patients suffering from neurological diseases.
Alternating Hemiplegia of Childhood (AHC), an extremely rare and severe neurological disease, often manifests quietly in infancy, bringing endless pain to affected children and their families. Its symptoms are particularly brutal, with sudden paralysis striking like a "time bomb," with each episode lasting from minutes to days. During these paralysis episodes, children often experience muscle dystonia, uncontrollable muscle stiffness, severe ocular movement issues, and developmental delays compared to their peers. Seizures, the disease's "lethal companion," constantly threaten the child's life. Currently, medical methods can only alleviate symptoms to a limited extent, making AHC a "fortress of stubborn disease" that remains unconquered and without a cure.
Researchers focused on the "culprits" of AHC—two common mutations D801N and E815K in the ATP1A3 gene. To investigate and test therapeutic solutions, Cathleen (Cat) Lutz developed a new mouse model. Previously, attempts to introduce these mutations into mice resulted in significant defects highly similar to human AHC and spontaneous early death in mice, providing an invaluable experimental model for this study.
Related Proteins
| Cat.No. # | Product Name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| ATP1A3-3253H | Recombinant Human ATP1A3 protein, His-tagged | E.coli | Human | His | 378-580 aa | |
| ATP1A3-366H | Recombinant Human ATP1A3 | Mammalian Cells | Human | His |
|
|
| ATP1A3-448R | Recombinant Rhesus monkey ATP1A3 Protein, His-tagged | Mammalian Cells | Rhesus macaque | His |
|
|
| ATP1A3-7024C | Recombinant Chicken ATP1A3 | Mammalian Cells | Chicken | His |
|
|
| ATP1A3-854R | Recombinant Rat ATP1A3 Protein | Mammalian Cells | Rat | His |
|
The research team tested two advanced next-generation technologies aimed at correcting mutations in genetically modified AHC mice. One was a well-known gene therapy that adds healthy copies of defective genes into the body, and the other was the emerging prime editing technique. Surprisingly, prime editing stood out in this "technical competition," demonstrating strong applicability. It acts like a precise "gene surgeon" capable of editing DNA bases and successfully repairing pathogenic gene mutations in up to 85% of brain cells. This directly resulted in a series of positive changes: normal protein function was restored in brain cells, motor skills improved significantly in mice, seizure occurrences reduced sharply, and the survival period was considerably extended.
Chinese Scientists Use AI-Driven Strategy for Rapid and Efficient Protein Evolution
DOI: 10.1016/j.cell.2025.06.014
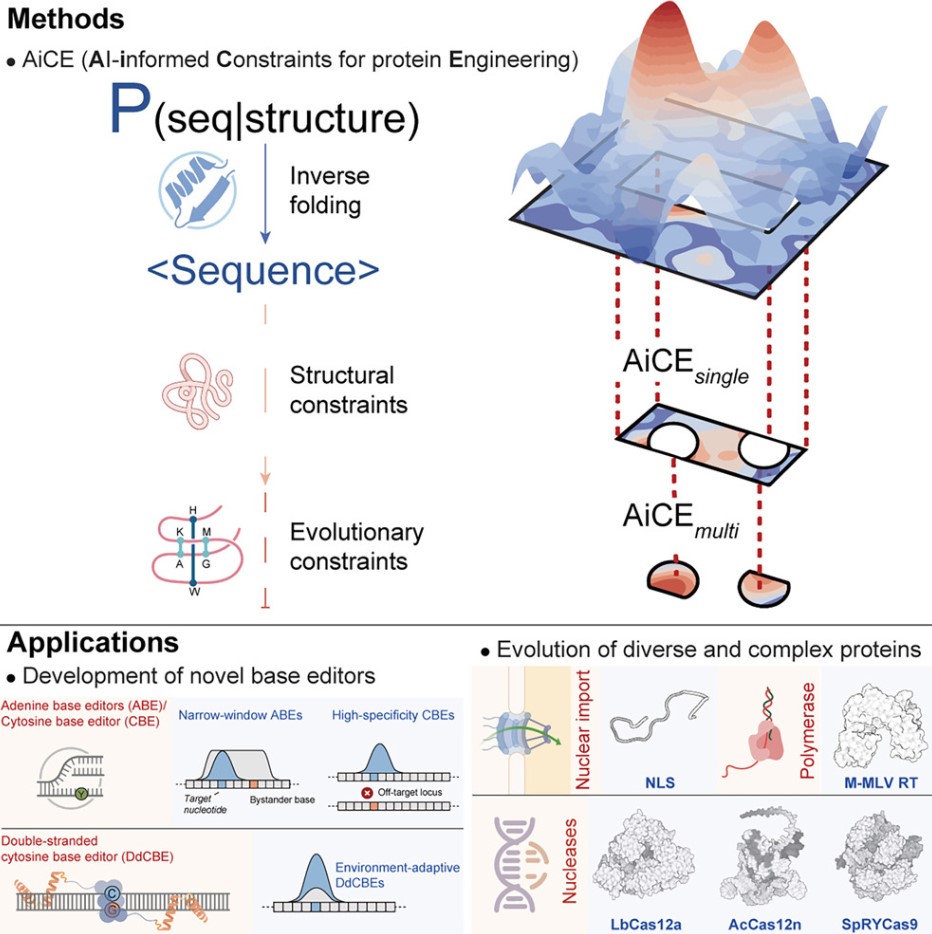
In a new study, a research team led by Professor Gao Caixia from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, developed a novel method that may revolutionize the protein engineering field. This new method, called AI-informed Constraints for protein Engineering (AiCE), integrates structural and evolutionary constraints into a general inverse folding model to achieve rapid and efficient protein evolution—without training specialized artificial intelligence (AI) models. This discovery addresses many challenges faced by traditional protein engineering techniques. The related research results were published in the journal Cell.
The ideal protein engineering strategy should achieve optimal performance with minimal effort. However, existing methods often face limitations in cost, efficiency, and scalability. Current AI-based protein engineering approaches tend to be computationally intensive, highlighting the need for more accessible and user-friendly alternatives that maintain prediction accuracy and promote broader applications in the research community.
In this study, researchers first developed AiCEsingle, a module for predicting high-fidelity (HF) single amino acid substitutions. This module improves prediction accuracy by extensively sampling an inverse folding model (an AI model that generates compatible amino acid sequences based on the protein's 3D structure) and integrating structural constraints.
Benchmark testing on 60 Deep Mutational Scanning (DMS) datasets showed that AiCEsingle's performance improved by 36% to 90% compared to other AI methods. Its effectiveness on complex proteins and protein-nucleic acid complexes was also validated. Notably, integrating structural constraints alone improved accuracy by 37%.
New Study Finds Compounds Activating Host Cell Integrated Stress Response Pathway Could Combat Various Viruses
DOI: 10.1016/j.cell.2025.06.024
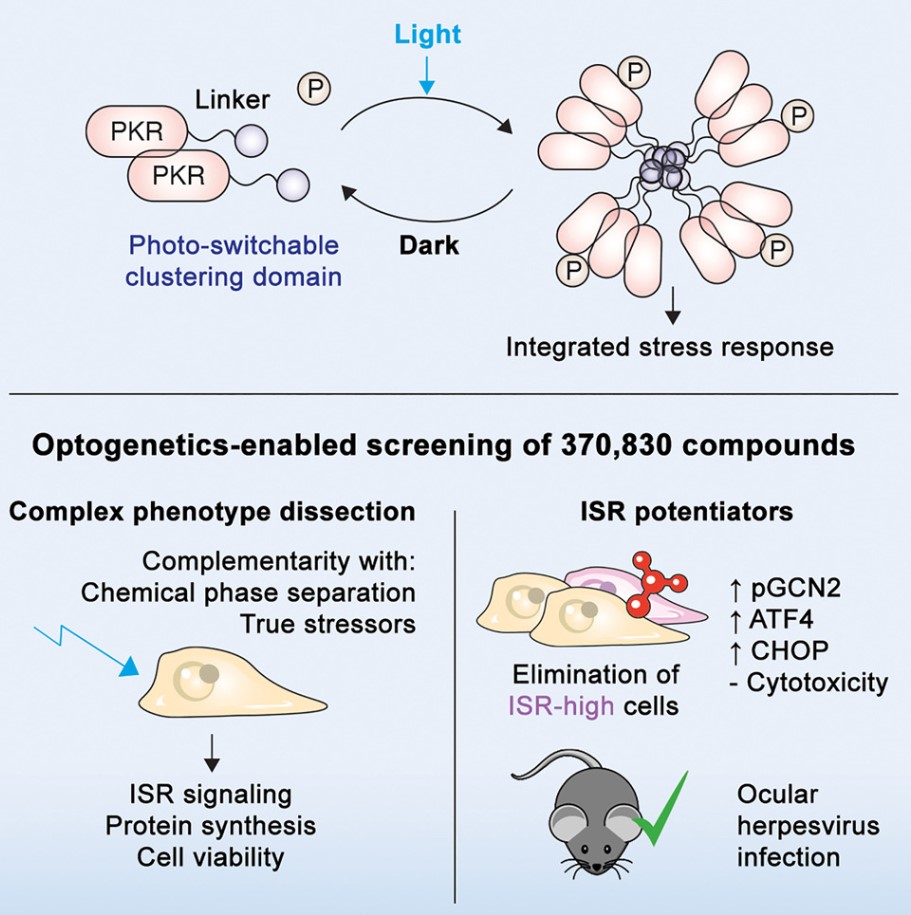
In a new study, researchers from MIT and other institutions discovered a class of compounds that can resist viral infections by activating a defense pathway within host cells. They believe these compounds hold promise as antiviral drugs effective not only against specific viruses but any type of virus. The related research findings were published in the journal Cell.
During screening of nearly 400,000 molecules, researchers identified compounds capable of activating the host cell defense system known as the integrated stress response pathway. In human cell experiments, they demonstrated these compounds help cells resist infections by respiratory syncytial virus (RSV), herpesvirus, and Zika virus. Their efficacy against herpesvirus infection was also shown in mouse models.
Researchers plan to further test the activity of these compounds against other viruses, aiming to develop them into clinical trial candidate drugs. James Collins, the study's co-corresponding author and Professor of Medical Engineering and Science at MIT’s Institute for Medical Engineering and Science, stated, "We are very excited about this study, which enables us to leverage the host cell's stress response to find methods for identifying and developing broad-spectrum antiviral drugs."
Discovery of Three New Types of Brain Waves Using Two Ultra-sensitive Optical Instruments
DOI: 10.1016/j.cell.2025.06.028
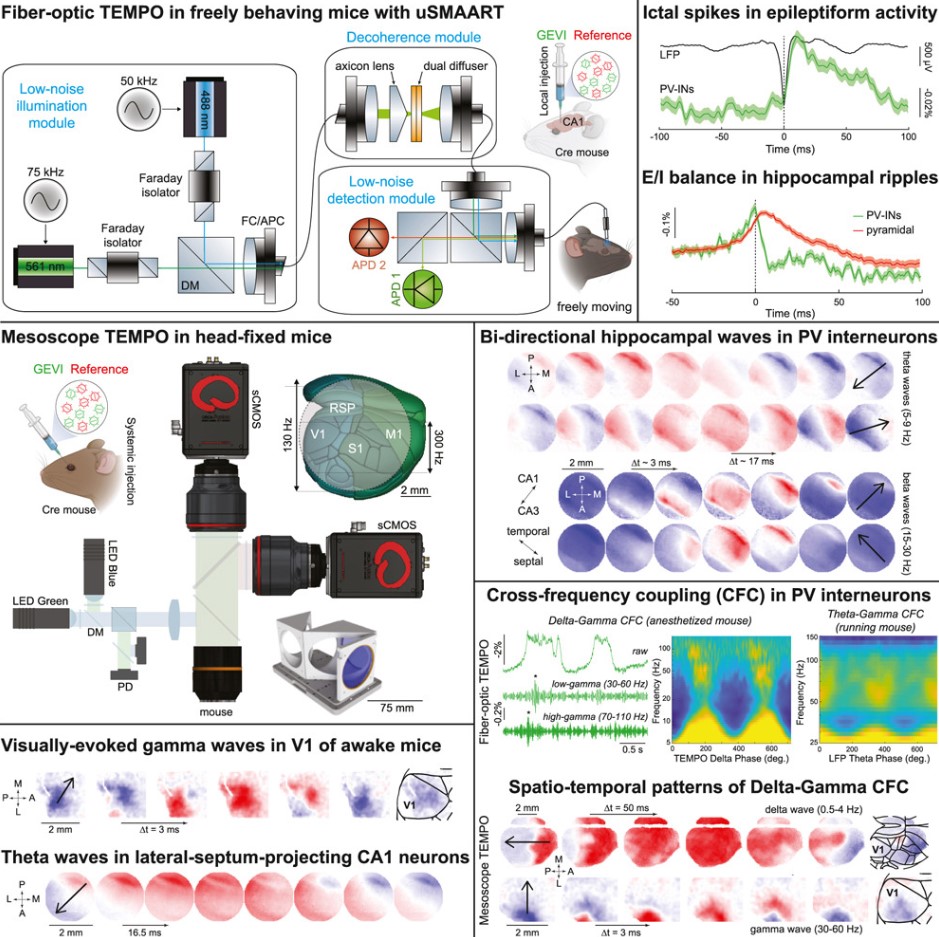
When electrical activity spreads in the brain, its motion resembles ripples in a pond. These "brain waves" were first observed in the 1920s. Now, in a new study, with the help of instruments and techniques developed by a Stanford University-led research team, their motion is clearer than ever.
This technology involves two ultra-sensitive optical instruments that can detect signals from gene-engineered proteins known as "voltage indicators," revealing the brain neuronal brain wave activity in mice. The related research findings were published in the journal Cell.
Although currently limited to animal research, this advancement has shown its potential. Using these instruments, the research team discovered three new types of brain waves with motion patterns never before observed.
Mark J. Schnitzer, the study's corresponding author and Professor of Biology and Applied Physics at Stanford University’s School of Humanities and Sciences, said, "We are gaining a broad perspective of brain wave propagation in the brain. We can observe multiple brain regions simultaneously and see brain waves sweep over the outermost layer of neural tissue—the cortex—in a cell-type-specific manner."
New Research Identifies Novel Receptor for AAV Virus Entry into Cells, Paving the Way for Safer and More Effective Gene Therapies
DOI: 10.1016/j.cell.2025.06.026
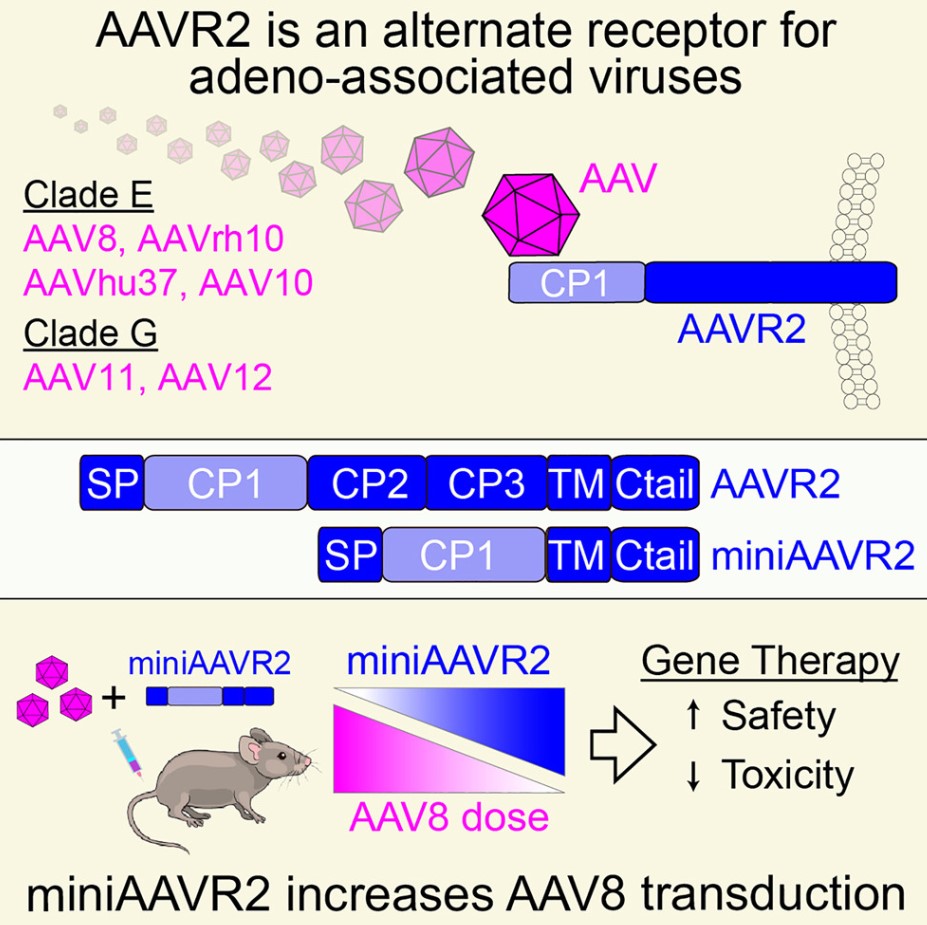
In new research from the University of Sydney in Australia and Peking University in China and other institutions, a milestone discovery was made that may offer safer and more effective gene therapies for a range of serious hereditary diseases, including Duchenne muscular dystrophy, Pompe disease, and hemophilia. The related research results were published in the journal Cell.
This study uncovered a previously unknown pathway for entering human cells—a receptor named AAVR2, utilized by gene therapy viruses to deliver therapeutic genes into cells. This newly discovered pathway may allow for lower doses of virus during treatment, reducing side effects and treatment costs while improving patient outcomes.
Gene therapy usually employs modified adeno-associated viruses (AAV) to deliver healthy genes into the body. Such treatments have the potential to change lives for patients, their families, and caregivers. However, they often require high doses of vectors to achieve therapeutic effects, which, in some cases, can provoke severe immune responses, lead to serious complications, or even death.
"We discovered that certain types of AAV can utilize the newly found AAVR2 receptor to enter cells, providing an alternative to previously known entry pathways. This discovery reveals a new route for gene delivery into cells. Regulating this pathway may make gene therapies safer, more economical, and more precise," said Dr. Bijay Dhungel, the study's first author and a researcher at the University of Sydney.
Related Products & Services
- Cytokines for Organoid Culture
- ADC Target Protein
- PROTAC Targets
- Cell and Gene Therapy
- Targets of CAR-T Cell Therapy
- Cancer Drug Targets
- Immune Checkpoint Proteins
- Protein Engineering Services
- Protein Interaction Service
- Protein Expression and Purification Services
- Drug Discovery Screening
- Protein Pathway Profiling
Reference
- Sousa AA, Terrey M, Sakai HA, Simmons CQ, Arystarkhova E, Morsci NS, Anderson LC, Xie J, Suri-Payer F, Laux LC, Roze E, Forlani S, Gao G, Frost S, Frost N, Sweadner KJ, George AL Jr, Lutz CM, Liu DR. In vivo prime editing rescues alternating hemiplegia of childhood in mice. Cell. 2025 Aug 7;188(16):4275-4294.e23. doi: 10.1016/j.cell.2025.06.038. Epub 2025 Jul 21. PMID: 40695277.
- Fei H, Li Y, Liu Y, Wei J, Chen A, Gao C. Advancing protein evolution with inverse folding models integrating structural and evolutionary constraints. Cell. 2025 Jul 3:S0092-8674(25)00680-4. doi: 10.1016/j.cell.2025.06.014. Epub ahead of print. PMID: 40628259.
- Wong F, Li A, Omori S, Lach RS, Nunez J, Ren Y, Brown SP, Singhal V, Lyda BR, Batjargal T, Dickson E, Rodrigues Reyes JR, Uruena Vargas JM, Wahane S, Kim H, Collins JJ, Wilson MZ. Optogenetics-enabled discovery of integrated stress response modulators. Cell. 2025 Jul 3:S0092-8674(25)00690-7. doi: 10.1016/j.cell.2025.06.024. Epub ahead of print. PMID: 40651473.
- Haziza S, Chrapkiewicz R, Zhang Y, Kruzhilin V, Li J, Li J, Delamare G, Swanson R, Buzsáki G, Kannan M, Vasan G, Lin MZ, Zeng H, Daigle TL, Schnitzer MJ. Imaging high-frequency voltage dynamics in multiple neuron classes of behaving mammals. Cell. 2025 Aug 7;188(16):4401-4423.e31. doi: 10.1016/j.cell.2025.06.028. Epub 2025 Jul 16. PMID: 40675148.
- Dhungel BP, Xu H, Nagarajah R, Vitale J, Wong ACH, Gokal D, Feng Y, Tabar MS, Metierre C, Parsania C, Song X, Wang G, Su XD, Bailey CG, Rasko JEJ. An alternate receptor for adeno-associated viruses. Cell. 2025 Jul 8:S0092-8674(25)00692-0. doi: 10.1016/j.cell.2025.06.026. Epub ahead of print. PMID: 40664211.
