Uncategorized Sunday, 2025/09/07
What makes the human brain unique? Cell: New study identifies two genes associated with human brain traits
DOI: 10.1016/j.cell.2025.06.037
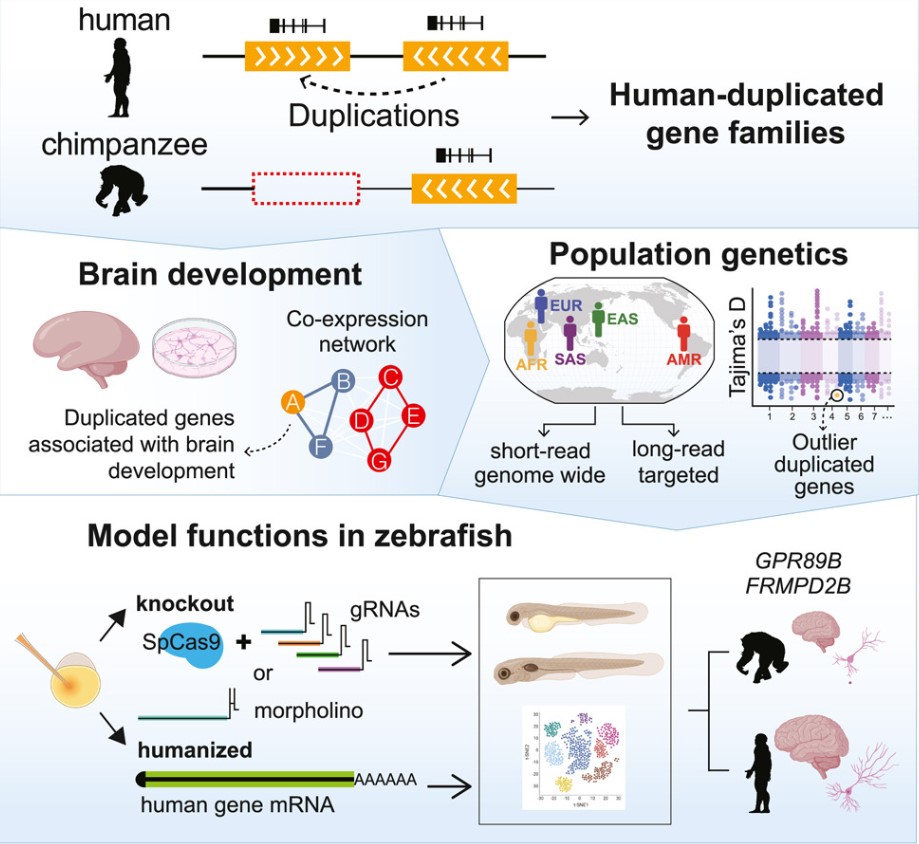
What makes the human brain stand out? A new study published in Cell has identified two genes linked to human brain traits and offers a roadmap for discovering more such genes. This research could help in understanding the functions and evolution of the human brain, as well as the roots of language disorders and autism.
Dennis and his colleagues used the telomere-to-telomere human genome to identify duplicated genes. They then filtered these based on sequences from the 1000 Genomes Project to find genes that are expressed in the brain, present in all humans, and conserved (i.e., they do not exhibit much variation among individuals).
They filtered out around 250 candidate gene families, and from these, they selected some for further study in the zebrafish animal model. By knocking out genes and introducing human duplicated genes into zebrafish, they demonstrated that at least two of these genes could contribute to human brain traits: a gene called GPR89B , which causes a slight increase in brain volume, and another gene called FRMPD2B , which alters synapse signaling.
Related Proteins
| Cat.No. # | Product Name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| GPR89B-1782R | Recombinant Rhesus Macaque GPR89B Protein, His (Fc)-Avi-tagged | HEK293 | Rhesus macaque | Avi&Fc&His |
|
|
| GPR89B-5276H | Recombinant Human GPR89B Protein, GST-tagged | Wheat Germ | Human | GST |
|
|
| GPR89B-5277H | Recombinant Human GPR89B Protein | Wheat Germ | Human | Non |
|
|
| FRMPD2B-285HCL | Recombinant Human FRMPD2L2 lysate | HEK293 | Human | Non |
|
New study identifies the world's first potential method to prevent life-threatening HTLV-1 virus infections
DOI: 10.1016/j.cell.2025.06.023
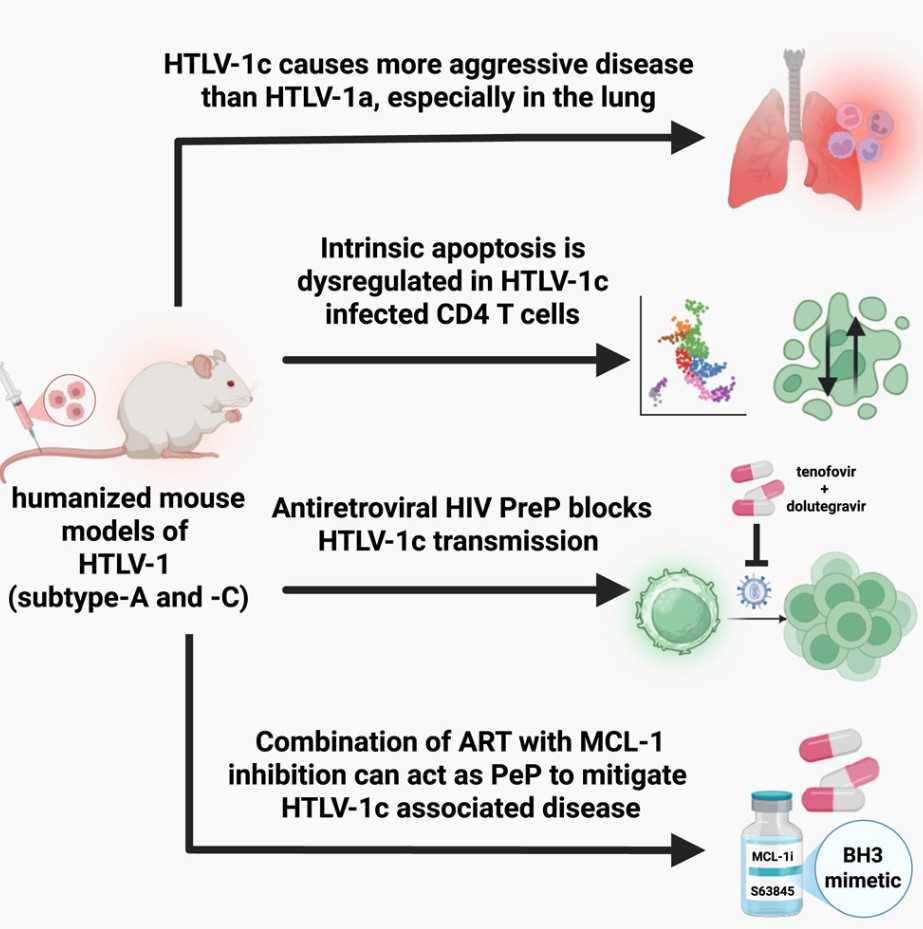
Human T-cell leukemia virus type 1 (HTLV-1) infects the same cell type as HIV, known as T cells, which are blood immune cells that help the body fight infections. A small portion of individuals infected with HTLV-1 develop severe diseases after a long period, such as adult T-cell leukemia and spinal cord inflammation. However, it remains a poorly understood disease, with no preventive treatments or cures available.
A groundbreaking new study might change this. Researchers from the United States and Australia have identified a new drug target that could clear HTLV-1-positive cells in infected individuals and prevent disease progression. They also discovered that existing HIV drugs could inhibit the spread of the HTLV-1 virus in mouse models. This finding could lead to the first treatment method to prevent the spread of this virus. The relevant research results are published in Cell.
Through a decade-long research effort, the team isolated the virus and developed the world's first humanized mouse model of HTLV-1 infection. This allowed them to study how the virus behaves in living organisms with a human-like immune system.
These mice were transplanted with human immune cells sensitive to HTLV-1 infection, including the unique Australian genotype strain HTLV-1c. International strains HTLV-1a and the Australian strain HTLV-1c induced leukemia and inflammatory lung disease in these mice with a human immune system. The mice were subsequently treated with tenofovir and dolutegravir—two antiviral therapies currently approved for suppressing HIV and preventing AIDS. The team found that these two drugs also effectively inhibit HTLV-1.
In another significant finding, the team discovered that when these mice were treated with a combination of HIV drugs and a therapy that inhibits the MCL-1 protein (known to help abnormal cells survive), human cells containing HTLV-1 could be selectively killed.
The team is now developing new methods targeting MCL-1 using precision RNA therapies and establishing a combinatorial treatment regimen ready for clinical testing. They believe this may provide a promising cure strategy for HTLV-1.
Related Proteins
| Cat.No. # | Product Name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| MCL1-781H |
Recombinant Human MCL1, His-tagged

|
E.coli | Human | His | 1-327aa | |
| Mcl1-185M | Recombinant Mouse Mcl1 protein, His-tagged | E.coli | Mouse | His | Phe2~Gly307 | |
| MCL1-3618R | Recombinant Rat MCL1 Protein | Mammalian Cells | Rat | His | ||
| MCL1-779H | Recombinant Human MCL1, His-tagged | E.coli | Human | His | 1-350aa | |
| Mcl1-780M | Recombinant Mouse Mcl1, GST-tagged | E.coli | Mouse | GST | 1-331aa | |
| MCL1-782H | Recombinant Human MCL1, GST-tagged | E.coli | Human | GST | 1-327aa |
New study suggests lapaquistat may treat Cryptosporidium infections
DOI: 10.1016/j.cell.2025.07.001
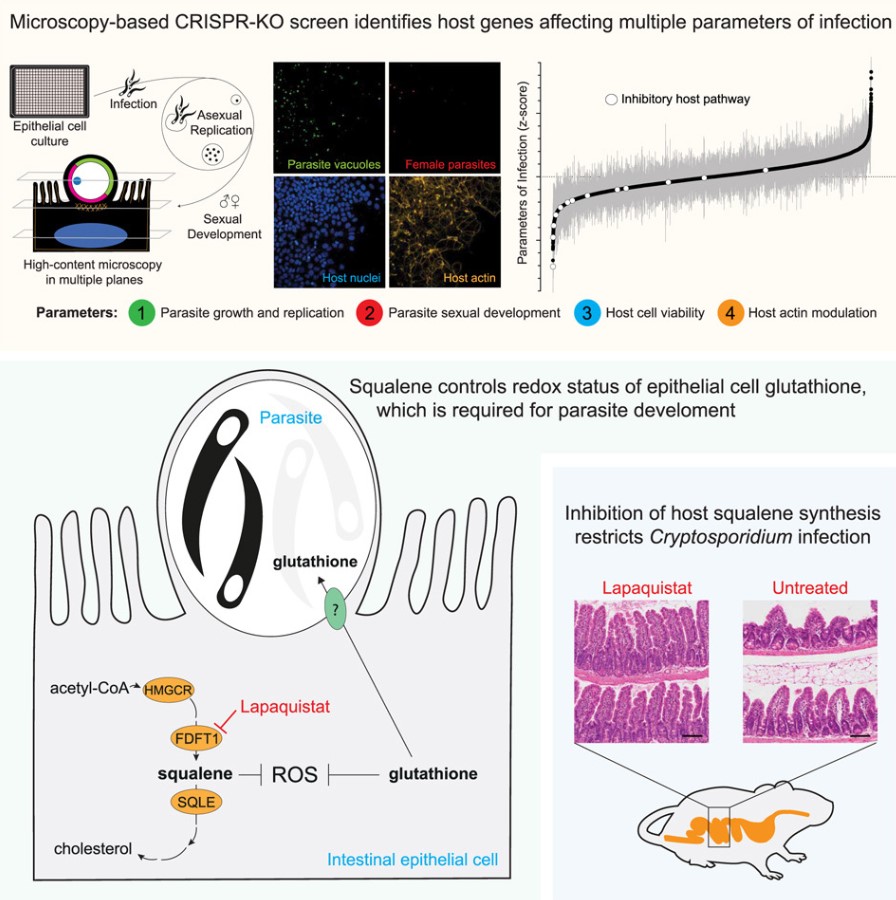
Cryptosporidium invades and multiplies within intestinal epithelial cells, causing severe diarrhea. This is especially dangerous for children in endemic areas and individuals with weakened immune systems. Despite its significant impact on global public health, no fully effective treatments currently exist. This parasitic invader has evolved the ability to survive in the human gut by manipulating biological pathways and hijacking host metabolism. In a new study, Adam and his team developed a method to map this complex survival roadmap and sever the Cryptosporidium's most crucial lifeline. The relevant findings are published in Cell.
Using CRISPR gene-editing technology, they conducted a genome-wide screening experiment that systematically switched off nearly 20,000 protein-coding genes in human intestinal cells. They then infected these cells with Cryptosporidium to observe how each gene influenced the parasite's survival.
Humans primarily manage oxidative stress through a molecule called glutathione, which is critical for limiting oxidative damage and is produced by almost all organisms. Surprisingly, the team found that while Cryptosporidium uses glutathione, it cannot produce it on its own. This makes the parasite reliant on the glutathione provided by intestinal cells and particularly vulnerable to oxidative stress.
Having identified this critical survival pathway, the team then sought ways to cut off this lifeline. While screening drugs targeting cholesterol synthesis, they discovered a previously abandoned drug showed promise because it directly blocked squalene production. When administered to mice infected with Cryptosporidium, this drug, named lapaquistat, alleviated the infection and prevented further intestinal damage.
AI + grammar model deciphers the dynamic code of cancer and brain development
DOI: 10.1016/j.cell.2025.06.048
In cancer research, whenever scientists want to test a new drug's effectiveness, it often requires lengthy cell culture and animal experiments, which are costly and time-consuming.
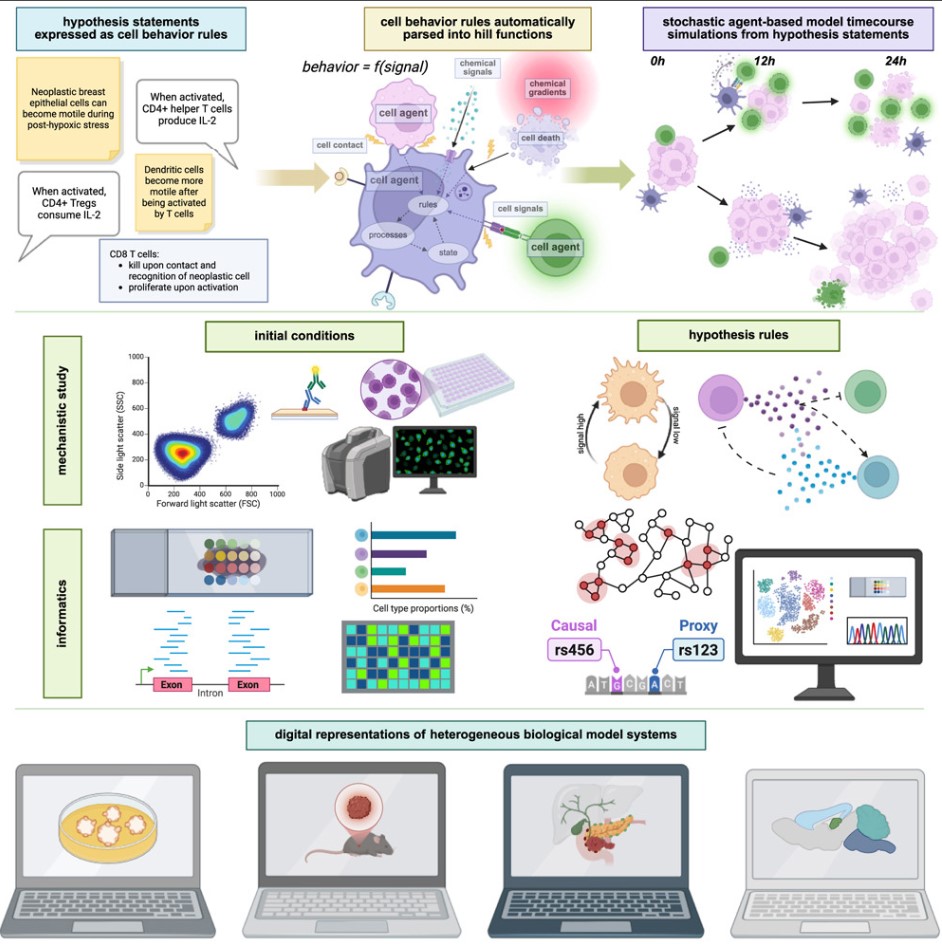
Now, a breakthrough study published in Cell might change this situation. A team from institutions including Indiana University and Johns Hopkins University School of Medicine has developed a "cell behavior grammar" that allows computers to interpret cell dynamics like human language, constructing a "digital twin" cell lab in a virtual environment to simulate complex processes from tumor growth to cortical formation.
Previously, computer modeling of cell behavior was a "technical task." Even creating a simple tumor growth model required researchers to master mathematical equations and programming code, often taking months to complete. The core breakthrough of the new research is establishing a "human-readable grammar" for cell behavior—like writing sentences in natural language. Scientists only need to input rules like "oxygen reduction makes tumor cells more motile," and the program can automatically translate this into a mathematical model that drives virtual cell behavior.
This grammar is built on an open-source software framework called PhysiCell, where "agents" act as "digital cells" following rules that map real cells' DNA, RNA instructions and can simulate complex behaviors such as cell division, migration, and interactions with other cells or drugs. For example, when modeling the tumor microenvironment, "agents" can change their state based on oxygen concentration: becoming more aggressive under low oxygen and reverting to quiescence under high oxygen.
"Past model coding took months; now, biologists can learn to build basic immunological models in two hours," said Paul Macklin, an engineering professor at Indiana University. Crucially, this grammar can directly connect with single-cell sequencing and spatial transcriptomics data, allowing virtual models to closely match the cellular distribution and functionality of real tissues.
Anti-cancer drugs "go beyond" to reverse Alzheimer's disease memory decline! Two old drugs combined hold promise
DOI: 10.1016/j.cell.2025.06.035
Alzheimer's disease (AD), the most common type of dementia, impacts millions globally. Characterized by amyloid-beta (Aβ) deposition, tau protein tangles, and abnormal neuron and glial cell function, it leads to progressive cognitive decline. However, research over past decades has only produced a few symptomatic drugs that cannot truly block or reverse disease progression.
Related Proteins
| Cat.No. # | Product Name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| MAPT-189H | Recombinant Human Tau-441 (244-372) | E.coli | Human | Non | 244-372 a.a. | |
| MAPT-01C | Recombinant Cynomolgus MAPT Protein, His tagged | E.coli | Cynomolgus | His | ||
| MAPT-02C | Recombinant Cynomolgus MAPT Protein, His tagged | HEK293 | Cynomolgus | His | ||
| Mapt-2928R |
Recombinant Rat Mapt protein(2-752aa), His-tagged
|
HEK293 | Rat | His | 2-752aa |
|
| Mapt-3222M |
Recombinant Mouse Mapt protein(2-733aa), His-tagged
|
HEK293 | Mouse | His | 2-733aa |
|
| MAPT-3533H |
Recombinant Human MAPT protein, His-tagged
|
HEK293 | Human | His | 1-441aa |
|
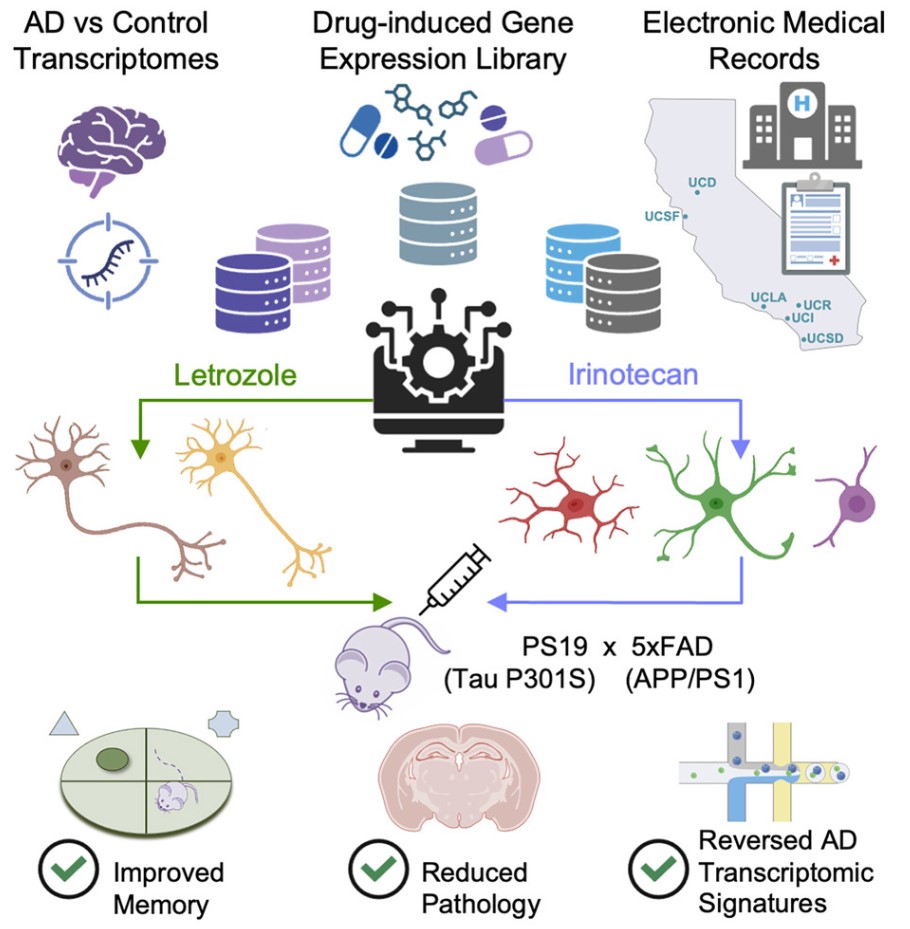
Recently, a breakthrough study published in Cell offers new hope: a team from the University of California, San Francisco, and the Gladstone Institutes discovered that a combination of two FDA-approved cancer drugs—letrozole and irinotecan—can precisely target abnormal gene expression in different cell types in the brains of AD patients, significantly reducing pathological damage and restoring memory in animal models. This "old drugs for new uses" discovery opens a new path for precise combination treatments for complex AD.
The study began by analyzing the molecular impact of AD on brain cells. The team integrated single-cell transcriptomics data from three public studies, covering brain samples from multiple AD patients and healthy controls, systematically analyzing gene expression changes in six key cell types (excitatory neurons, inhibitory neurons, microglia, astrocytes, oligodendrocytes, and oligodendrocyte precursor cells). The results revealed that AD not only alters the gene activity of neurons, but also involves inflammatory responses and metabolic disorders in glial cells, with distinct abnormal gene characteristics in different cells.
Based on these cell-specific gene "abnormal maps," researchers screened 1,300 FDA-approved drugs—comparing gene expression changes induced by each drug in human cells to identify candidates that could "reverse" AD's abnormal traits. Ultimately, 86 drugs were initially identified, and 25 could affect multiple cell types.
For further validation, the team reviewed 1.4 million electronic medical records of individuals over 65 from the University of California system, analyzing these drugs' association with AD onset risk. Results showed that five drugs were significantly associated with reduced AD risk, notably letrozole (commonly used for breast cancer treatment) and irinotecan (commonly used for colorectal cancer treatment): populations taking letrozole had significantly reduced AD relative risk, and those using irinotecan showed even lower risk. Notably, letrozole mainly targets neurons' abnormal gene expression, while irinotecan focuses on glial cells, making them highly complementary.
Related Products & Services
- Cytokines for Organoid Culture
- ADC Target Protein
- PROTAC Targets
- Cell and Gene Therapy
- Targets of CAR-T Cell Therapy
- Cancer Drug Targets
- Immune Checkpoint Proteins
- Protein Engineering Services
- Protein Interaction Service
- Protein Expression and Purification Services
- Drug Discovery Screening
- Protein Pathway Profiling
Reference
- Soto DC, Uribe-Salazar JM, Kaya G, Valdarrago R, Sekar A, Haghani NK, Hino K, La G, Mariano NAF, Ingamells C, Baraban A, Jamal Z, Turner TN, Green ED, Simó S, Quon G, Andrés AM, Dennis MY. Human-specific gene expansions contribute to brain evolution. Cell. 2025 Jul 18:S0092-8674(25)00739-1. doi: 10.1016/j.cell.2025.06.037. Epub ahead of print. PMID: 40695280; PMCID: PMC12313184.
- Cooney JP, Hirons A, Jansz N, Allison CC, Hickey P, Teh CE, Tan T, Dagley LF, Yousef J, Yurick D, Khoury G, Preston SP, Arandjelovic P, Davidson KC, Williams LJ, Bader SM, Wang L, Bhandari R, Mackiewicz L, Dayton M, Clow W, Faulkner GJ, Gray DH, Einsiedel L, Purcell DFJ, Doerflinger M, Pellegrini M. Combination antiretroviral therapy and MCL-1 inhibition mitigate HTLV-1 infection in vivo. Cell. 2025 Jul 1:S0092-8674(25)00689-0. doi: 10.1016/j.cell.2025.06.023. Epub ahead of print. PMID: 40645177.
- Marzook NB, Song OR, Baumgärtel L, Bernitz N, Mkandawire TT, Watson LC, Nunes V, Warchal S, MacRae JI, Howell M, Sateriale A. The essential host genome for Cryptosporidium survival exposes metabolic dependencies that can be leveraged for treatment. Cell. 2025 Jul 17:S0092-8674(25)00751-2. doi: 10.1016/j.cell.2025.07.001. Epub ahead of print. PMID: 40706591.
- Johnson JAI, Bergman DR, Rocha HL, Zhou DL, Cramer E, Mclean IC, Dance YW, Booth M, Nicholas Z, Lopez-Vidal T, Deshpande A, Heiland R, Bucher E, Shojaeian F, Dunworth M, Forjaz A, Getz M, Godet I, Kurtoglu F, Lyman M, Metzcar J, Mitchell JT, Raddatz A, Solorzano J, Sundus A, Wang Y, DeNardo DG, Ewald AJ, Gilkes DM, Kagohara LT, Kiemen AL, Thompson ED, Wirtz D, Wood LD, Wu PH, Zaidi N, Zheng L, Zimmerman JW, Phillip JM, Jaffee EM, Gray JW, Coussens LM, Chang YH, Heiser LM, Stein-O'Brien GL, Fertig EJ, Macklin P. Human interpretable grammar encodes multicellular systems biology models to democratize virtual cell laboratories. Cell. 2025 Jul 25:S0092-8674(25)00750-0. doi: 10.1016/j.cell.2025.06.048. Epub ahead of print. PMID: 40713951.
- Li Y, Pereda Serras C, Blumenfeld J, Xie M, Hao Y, Deng E, Chun YY, Holtzman J, An A, Yoon SY, Tang X, Rao A, Woldemariam S, Tang A, Zhang A, Simms J, Lo I, Oskotsky T, Keiser MJ, Huang Y, Sirota M. Cell-type-directed network-correcting combination therapy for Alzheimer's disease. Cell. 2025 Jul 15:S0092-8674(25)00737-8. doi: 10.1016/j.cell.2025.06.035. Epub ahead of print. PMID: 40695276; PMCID: PMC12313259.
