Uncategorized Tuesday, 2025/05/13
Alzheimer’s disease (AD) is the most common neurodegenerative dementia, and accurate pathological staging is crucial for treatment trials and prognosis. The classification of AD as proposed by the National Institute on Aging and Alzheimer’s Association (NIA-AA) relies on biomarkers of Aβ protein pathology (A) and phosphorylated tau (T), yet there remains a need to develop more cost-effective and sensitive CSF biomarkers for early diagnosis and pathological staging of AD.
Today, we share high-level articles published in Nature Medicine, Cell, Nature Human Behavio, Neuron, and Cell Reports Medicine, elucidating how SomaScan technology can be leveraged to perform proteomic analysis of cerebrospinal fluid (CSF) to discover AD biomarkers.
Case 1: CSF Proteomics Discovers New Predictive Biomarkers for Cognitive Decline in Alzheimer’s Independent of Aβ/Tau
On March 31, 2025, a global collaboration involving Stanford University, the Hong Kong Neurodegenerative Disease Research Centre, and 35 other institutions published an article titled "A cerebrospinal fluid synaptic protein biomarker for prediction of cognitive resilience versus decline in Alzheimer's disease" in Nature Medicine. Utilizing SomaScan technology, the study conducted proteomic analysis on 3,397 CSF and 13,401 plasma samples from various cohorts, employing machine learning methods to identify the YWHAG:NPTX2 synaptic protein ratio. This ratio (where YWHAG interacts with tau protein to regulate its aggregation and phosphorylation; NPTX2 is a secreted synaptic protein essential for synaptic transmission, plasticity, and neural circuit homeostasis) better explains the variability in cognitive impairment severity than gold standard biomarkers like pTau181:Aβ42 and tau PET. The YWHAG:NPTX2 ratio’s predictive ability for AD cognitive impairment was partially reproduced in blood samples, offering a more sensitive and convenient biomarker for clinical application.

Fig1. The CSF YWHAG:NPTX2 ratio explains a substantial proportion of variance in CI beyond amyloid and tau in AD.
Experimental Design: The study integrated 3,397 CSF samples and 13,401 plasma samples from 6 major prospective Alzheimer's case-control cohorts. Through large-scale proteomic screening, the YWHAG to NPTX2 synaptic protein ratio was found to be significantly associated with cognitive impairment. The robustness of these protein biomarkers was validated using high-accuracy mass spectrometry technologies (LC-MS/MS and TMT-MS) in the BioFINDER2 and Kuopio cohorts, ensuring cross-platform reproducibility. Plasma proteomics screening was used to construct molecular biomarker combinations predicting cognitive impairment (including synaptic proteins like CPLX2 and NPTXR). The strong correlation between plasma-derived protein biomarkers and CSF YWHAG:NPTX2 was verified, validating the feasibility of blood tests as a substitute for CSF.
| Cat.No. # | Product Name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| MAPT-189H | Recombinant Human Tau-441 (244-372) | E.coli | Human | Non | 244-372 a.a. | |
| MAPT-01C | Recombinant Cynomolgus MAPT Protein, His tagged | E.coli | Cynomolgus | His | ||
| MAPT-02C | Recombinant Cynomolgus MAPT Protein, His tagged | HEK293 | Cynomolgus | His | ||
| YWHAG-100H | Recombinant Human YWHAG , His-tagged | E.coli | Human | His | 2-247 a.a. |
|
| YWHAG-12H | Recombinant Human YWHAG | E.coli | Human | Non | 1-247 a.a. |
|
| NPTX2-1349H | Recombinant Human NPTX2, GST-tagged | E.coli | Human | GST | N-term-350aa | |
| NPTX2-487H | Recombinant Human NPTX2 Protein, hFc-tagged | HEK293 | Human | Fc | 1-431 aa |
|
Case 2: CSF Proteomics Identifies Early Changes in Autosomal Dominant Alzheimer's Disease (ADAD)
On September 19, 2024, a team led by Carlos Cruchaga from Washington University published an article titled "CSF proteomics identifies early changes in autosomal dominant Alzheimer’s disease" in Cell. Through high-throughput CSF proteomics (using SomaScan technology), the study revealed early proteomic changes in ADAD, identified early ADAD biomarkers, and developed a robust predictive model for ADAD. The research identified 137 CSF proteins exhibiting distinct trajectories between MC and NC groups, changing 15-20 years before disease onset, involving pathways such as neuronal death and immune response. The study also discovered six subgroups of proteins that more effectively differentiate MCs and NCs, showing superior predictive power compared to conventional biomarkers. These findings emphasize the similarities and differences between ADAD and sporadic AD, providing personalized medical possibilities for individuals carrying ADAD gene mutations.
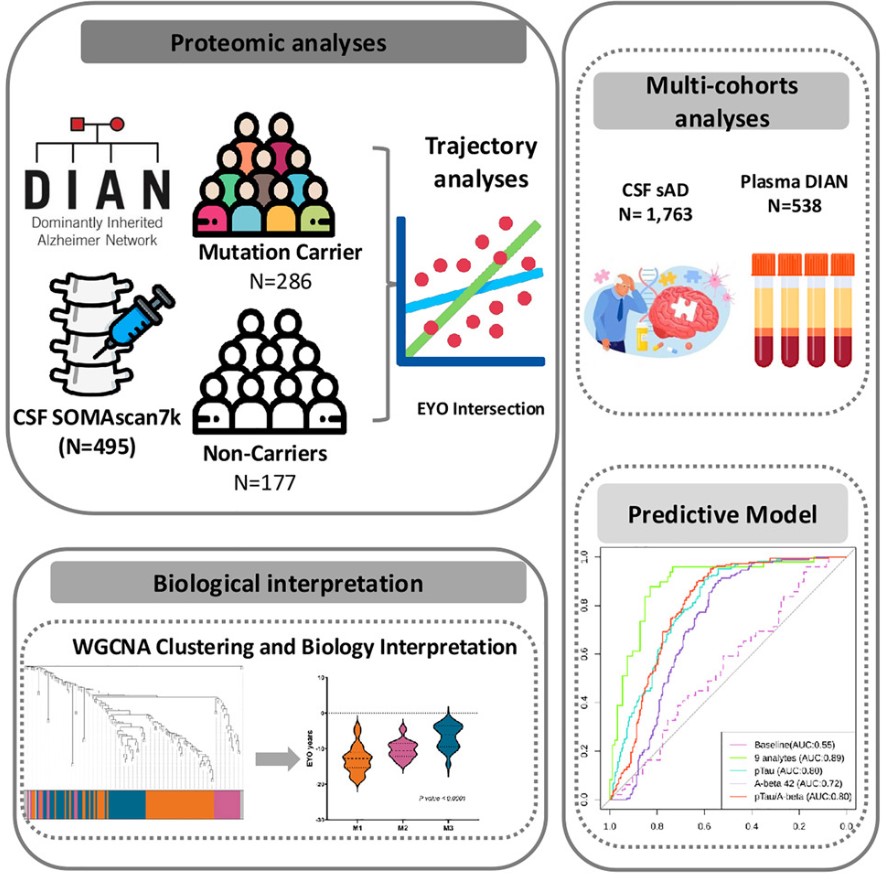
Fig2. Graphical abstract
Experimental Design: The study included ADAD and sporadic AD cohorts. The ADAD cohort used the SomaScan 7K panel to analyze CSF and plasma proteomes of DIAN participants, with CSF samples from 286 mutation carriers (MCs) and 177 non-carriers (NCs), and plasma samples from 325 MCs and 213 NCs. A total of 448 participants had both CSF and plasma proteome data. The CSF samples included 212 (74%) PSEN1 mutation carriers, 23 (8%) PSEN2 mutation carriers, and 51 (18%) APP mutation carriers, with 102 symptomatic MCs (36%) and 184 asymptomatic MCs (64%); NCs were recruited from MCs' family members. The sporadic AD cohort served as a validation cohort with 1,763 individual samples from four different research centers.
| Cat.No. # | Product Name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| PSEN1-2004H | Recombinant Human PSEN1 Protein, His-tagged | E.coli | Human | His | 254-377aa | |
| PSEN1-23HFL | Recombinant Full Length Human presenilin 1 Protein, Flag tagged | HEK293 | Human | Flag | Full L. 1-467aa |
|
| PSEN2-28H | Recombinant Human presenilin 2 (Alzheimer disease 4) Protein, His-tagged | E.coli | Human | His | 305-384aa | |
| PSEN2-28527TH | Recombinant Human PSEN2 | Wheat Germ | Human | Non | 86 amino acids |
|
| APP-3646H | Recombinant Human APP protein, His-tagged | E.coli | Human | His | 653-751 aa | |
| APP-3850H | Recombinant Human APP, His & GST-tagged | E.coli | Human | GST&His | Asp672-Ala713 |
Case 3: CSF Proteomics Identifies Biomarkers for Amyloid and Tau PET Staging
On July 10, 2024, a team led by Professor Yujin Tai from the Department of Neurology, Huashan Hospital, Fudan University, in collaboration with Fudan University's Institute of Brain-Inspired Intelligence, published an article in Nature Human Behavior titled "Multiplex cerebrospinal fluid proteomics identifies biomarkers for diagnosis and prediction of Alzheimer’s disease." The study revealed AD biomarkers like YWHAG, which performed best in biologically defined AD diagnosis. Four-protein (YWHAG, SMOC1, PIGR, TMOD2) and five-protein (ACHE, YWHAG, PCSK1, MMP10, IRF1) combinations reached diagnostic accuracies of 0.987 and 0.975 respectively, providing new insights for clinical trials targeting different AD pathological mechanisms and showing promise in clinical applications and future research.
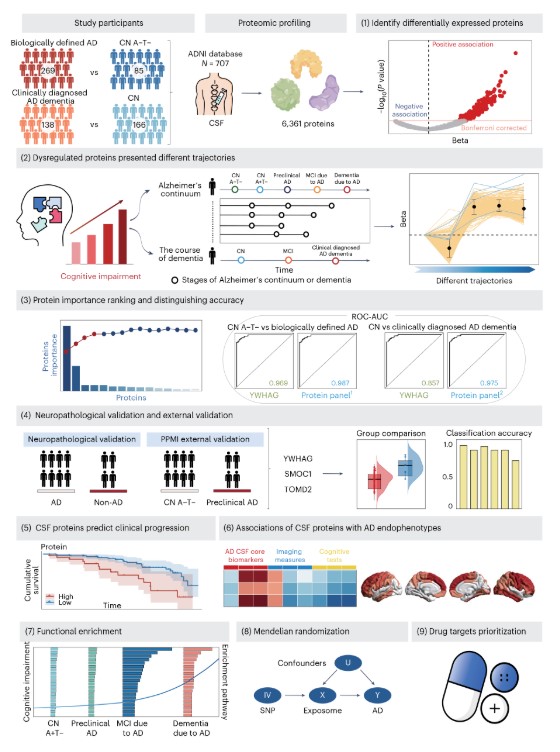
Fig3. Study overview
Experimental Design: The study involved 707 participants, including cognitively normal (CN), mild cognitive impairment (MCI), and clinically diagnosed AD dementia patients. Through differential protein expression analysis using the largest high-throughput CSF proteomics dataset (SomaScan platform, 6,361 proteins) to date, protein dynamic change trajectories and importance rankings were analyzed for modeling accuracy differentiation. Pathological validation through autopsy and external validation with the PPMI (Parkinson’s Progression Markers Initiative) were conducted. Additionally, the authors evaluated the performance of these proteins in predicting clinical progression risks and linked these proteins to AD endophenotypes. Dysfunctional proteins underwent functional enrichment analysis, Mendelian randomization, and drug-like property analysis.
Case 4: Multi-Cohort CSF Proteomics Identifies Robust Molecular Signatures Across the Alzheimer Disease Continuum
On March 14, 2025, Carlos Cruchaga’s team at the Knight Alzheimer’s Disease Research Center at Washington University published an article in Neuron titled "Multi-cohort cerebrospinal fluid proteomics identifies robust molecular signatures across the Alzheimer disease continuum." Using SomaScan technology, over 6,000 proteins in CSF were targeted for proteomic analysis, identifying 2,173 dysregulated proteins in AD, enriched in pathways including neuronal death, apoptosis, tau phosphorylation, microglial dysregulation, lysosomal dysfunction, brain plasticity, longevity, and microglia-neuron crosstalk. Machine learning was used to create and validate highly accurate and reproducible models (AUC> 0.90) predicting AD biomarker positivity and clinical states, also identifying those who will convert to AD.
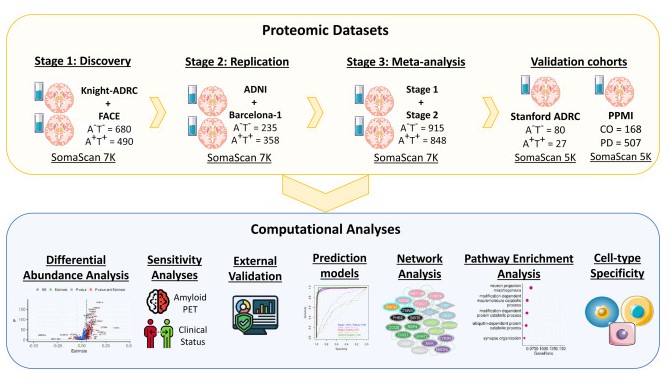
Fig4. Schematic of experimental and analytical workflow
Experimental Design: The study measured 7,029 proteins in CSF of 2,286 participants (from Knight ADRC, FACE, ADNI, and Barcelona-1 cohorts) using SomaLogic 7K. Machine learning determined the minimal protein set predicting biomarker status, analyzing whether these proteins could also predict clinical and amyloid imaging states. Longitudinal clinical data was utilized to determine if the identified proteomic features could also predict progression from control to AD. Specificity analysis was performed using proteomic data from frontotemporal dementia (FTD), Lewy body dementia (DLB), and Parkinson’s disease (PD). Lastly, differential protein abundance in different AT groups (A−T− vs A+T−, A+T− vs A+T+) was comprehensively examined to reveal distinct protein pseudo-trajectories across the AD spectrum, based on cross-sectional data estimating protein longitudinal trajectories. Group-specific pathway enrichment analyses were conducted to describe biological processes impacted at different AD continuum stages.
Case 5: CSF Proteomics Identifies Biomarkers for Amyloid and Tau PET Stages
On March 20, 2025, the team led by Jiajianping at Xuanwu Hospital, Capital Medical University, published an article in Cell Reports Medicine titled "Cerebrospinal fluid proteomics identification of biomarkers for amyloid and tau PET stages." Using SomaScan technology, over 6,000 proteins in the CSF were targeted for proteomic analysis, combined with machine learning to identify candidate biomarkers separating amyloid PET and tau PET stages, successfully developing two protein panels distinguishing early and late disease stages as defined by PET imaging. These models outperformed established CSF AD biomarkers (i.e., Aβ42 and p-tau181) in stage differentiation and were linked to dementia progression and cognitive decline over subsequent decades. Proteins in the early stage were rich in synaptic damage and compensatory processes, while late-stage proteins were associated with metabolic dysfunction pathways.
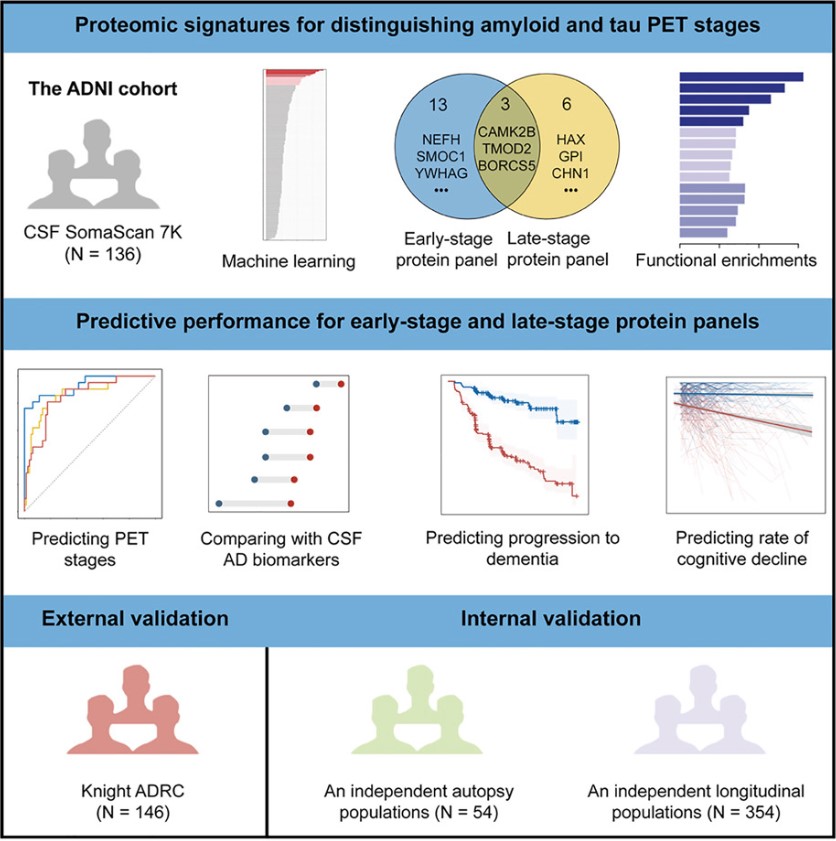
Fig5. Graphical abstract
Experimental Design: The primary objective was to identify CSF proteins exhibiting changes in early and late AD as assessed by amyloid PET and tau PET. The main cohort included 136 non-dementia individuals, with a mean follow-up of 4.4 years. During this time, 14 participants (11.80%) converted from amyloid PET negative (A−) to positive (A+), and 31 participants (22.80%) progressed to dementia. For validation, an independent cohort of 54 participants was analyzed, including 11 with no/low AD neuropathological changes (ADNC) and 43 with moderate/high ADNC. Additionally, a longitudinal cohort of 354 non-dementia individuals was assessed for clinical progression over an average of 3.70 years. Analysis included 6,164 proteins in total. Established AD biomarkers, CSF Aβ42, p-tau181, t-tau, and p-tau181/Aβ42 ratio, were used for comparison. Machine learning techniques determined protein subsets that accurately distinguished early and late AD stages. Additional analysis explored the relationship between these proteins and dementia progression and cognitive decline, using external and internal cohorts for validation.
Summary
Cerebrospinal fluid (CSF) is a valuable source for understanding changes occurring in the brain during neurodegenerative diseases, providing insights into their underlying pathologies. Systematic exploration of the CSF proteome holds the potential to identify new biomarkers reflecting the multifaceted pathophysiology of AD. Current studies leveraging CSF and plasma proteomics to identify AD-related proteins and pathways are numerous. Despite their informativeness, they often investigate a limited number of proteins, as seen with Olink and mass spectrometry, hampering the identification of AD-associated proteins, biomarkers, and effective disease targets. Currently, Creative BioMart provides a list of related proteins and biomarkers for researcher to accelerate their program.
Related Products & Services
- Neurological Disorders Targets Adhesion Molecules
- Blood-brain Barrier Permeability
- Calcium-binding Proteins and Related Molecules
- Neurodegenerative Disease
- Neurotransmitter Associated Enzymes
- Neurotransmitter Receptors, Transporters, and Ion Channels
- Neurotrophic Factors and Receptors
- Synaptic Proteins and Receptors
- Protein Engineering Services
- Protein Interaction Service
- Protein Expression and Purification Services
- Drug Discovery Screening
- Protein Pathway Profiling
Reference
- Oh, H. S., Urey, D. Y., Karlsson, et.al (2025). A cerebrospinal fluid synaptic protein biomarker for prediction of cognitive resilience versus decline in Alzheimer’s disease. Nature Medicine, 1-12. https://doi.org/10.1038/s41591-025-03565-2
- Shen, YuanyuanNoble, James M. et al. CSF proteomics identifies early changes in autosomal dominant Alzheimer’s disease. Cell, Volume 187, Issue 22, 6309 - 6326.e15
- Guo, Y., Chen, S., You, J., et al(2024). Multiplex cerebrospinal fluid proteomics identifies biomarkers for diagnosis and prediction of Alzheimer’s disease. Nature Human Behaviour, 8(10), 2047-2066. https://doi.org/10.1038/s41562-024-01924-6
- Ali, Muhammad et al. Multi-cohort cerebrospinal fluid proteomics identifies robust molecular signatures across the Alzheimer disease continuum. Neuron, Volume 0, Issue 0
- Wang, Zhibo et al. Cerebrospinal fluid proteomics identification of biomarkers for amyloid and tau PET stages. Cell Reports Medicine, Volume 6, Issue 4, 102031
