In eukaryotes, the highly folded structure is necessary to package chromatin into the nucleus, but such dense chromatin blocks gene transcription, DNA replication, and damage repair at the corresponding chromatin sites. Therefore, eukaryotes evolved to produce a set of chromatin remodeling enzymes and some related factors, which regulate the structure of chromatin by regulating the assembly, disassembly, and relocalization of nucleosomes. There are proteins that can exploit ATP hydrolysis to produce energy to drive nucleosome sliding, or mediated replacement between histone variants and classic histones in nucleosomes. These proteins are ATP-dependent chromatin remodeling factors, which typically consist of multiple subunits to form a larger chromatin remodeling complex. Different chromatin remodeling factors are located on specific nucleosomes at specific times, affecting gene transcriptional activity by altering chromatin structure, thus ensuring the proper functioning of various biological processes within the cell.
Classification of Chromatin Remodeling Factors
Chromatin remodeling factors can be roughly divided into four families, SWI/SNF, ISWI, CHD, and INO80, according to the different functional domains they contain. Different chromatin remodeling factors share similarities in protein structure and enzyme activity, while each has its specificity. The chromatin remodeling complex relies on the energy generated by the hydrolysis of ATP to perform the remodeling function, and its core subunit is ATPase catalytic subunit. The classification of remodeling complex subfamilies is classified according to the structural characteristics of its catalytic subunit, which can be classified into four types, namely SWI/SNF, ISWI, CHD, and INO80/SWR1. The ATPase catalytic subunits of diverse subfamilies have similar functional domains, but also have specific amino acid sequences and domains.
Table 1 Classification and subunit composition of chromatin remodeling complexes
| Superfamily | Complex (Number of Subunits)-Yeast | Catalytic Subunit-Yeast | Complex (Number of Subunits)-Human | Catalytic Subunit-Human |
|---|---|---|---|---|
| SWI/SNF | SWI/SNF (12) | Swi2/Snf2 | BAF (10) | hBrm or Brg1 |
| RSC (17) | Sth1 | PBAF (12) | Brg1 | |
| ISWI | ISWI (2) | ISWI1 or ISWI2 | ACF (2) | Snf2H |
| ISWIb (3) | CHRAC (4) | |||
| ISWI2 (2) | NoRC (2) | |||
| RSF (2) | ||||
| WICH (2) | ||||
| NURF (3) | Snf2L | |||
| CHD | CHD1 (1) | Chd1 | CHD1 (1) | Chd1 |
| CHD2 (1) | Chd2 | |||
| NuRD (7) | Chd3/Chd4 | |||
| INO80 | yINO8 (15) | Ino80 | INO80 (15) | hIno80 |
| SWR1 (16) | Swr1 | SRCAP (9) | SRCAP | |
| TRRAP/Tip60 (16) | P400 |
Subunit Composition of Chromatin Remodeling Complex
Except for CHD1 and CHD2 chromatin remodeling enzymes that can exert their remodeling functions alone, most remodeling enzymes perform remodeling functions in the form of multi-subunit complexes in vivo, hence they are termed as chromatin remodeling complexes. Each remodeling complex contains a different number of subunits. For example, the ISWI family complex has only a few subunits, while the Ino80 and SWI2 complexes in the Ino80 family have up to a dozen subunits. Although the role of most subunits in chromatin remodeling complexes is not well understood, it is speculated that they play important roles in specific site recognition, binding of specific proteins, stabilization of complex structures, and regulation of enzymatic activity.
Functional Mechanisms of Chromatin Remodeling Factor
The chromatin remodeling complex allows nucleosomes to slide along the DNA without unwinding the double-stranded DNA. The DNA bulge created by these complexes will alter the interaction between histone octamer and DNA, resulting in changes in the relative position of DNA on the histone surface.
Most nucleosomes in chromatin are composed of four classic histones (H2A/H2B/H3/H4), but some of the classic histones in nucleosomes can be replaced by histone variants. Nucleosomes containing histone variants are specially labeled on chromatin, and the specific structural characteristics of different variants cause certain changes in chromatin structure, thereby mediating changes in corresponding cellular functions, such as gene transcription regulation and DNA damage repair. It has been found that some chromatin remodeling complexes are the performers of the replacement of histone variants into (or out of) nucleosomes. For example, Swr1 catalyzes the substitution of the heterodimer H2AZ-H2B with the classic H2A-H2B dimer in nucleosomes in yeast. Besides H2AZ, there are many intracellular histone variants such as H3.3, H2AX, CENP-A.
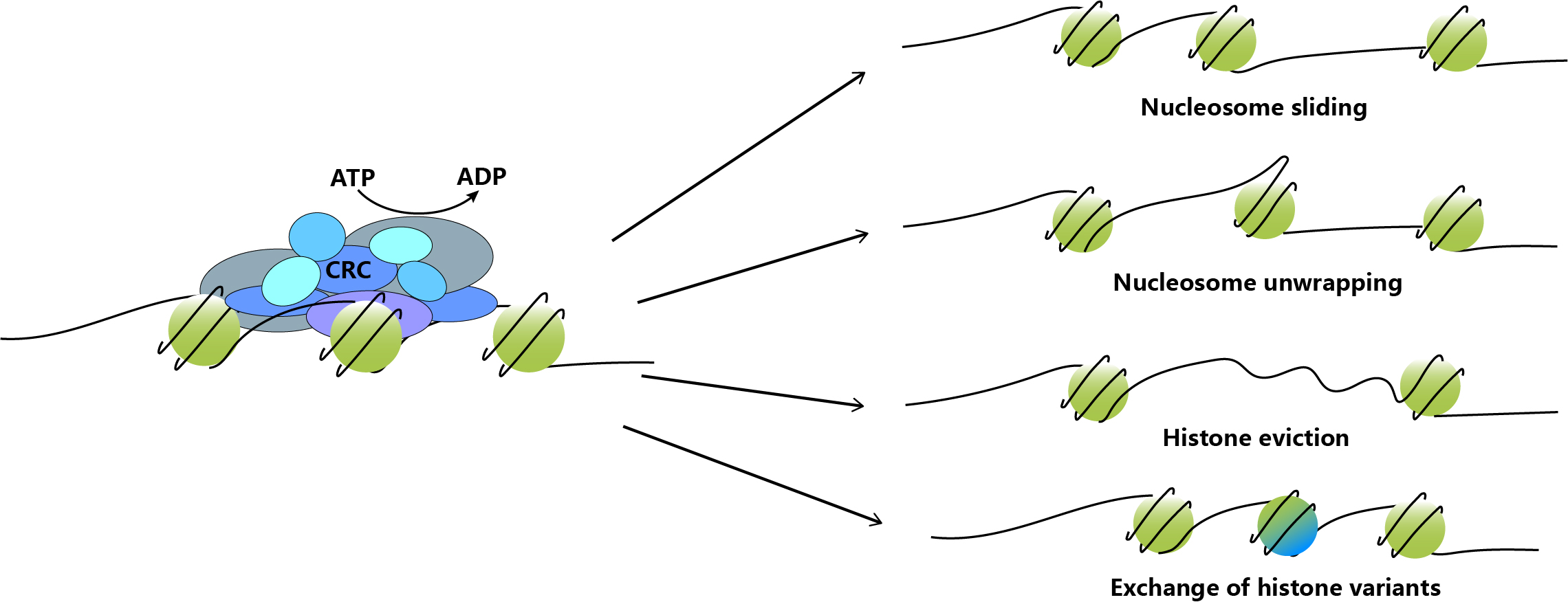
Figure 1. Schematic of the ATP-dependent nucleosome accessibility regulation.
Chromatin Remodeling Factor-mediated Gene Transcription Regulation
Chromatin structure in cells is under dynamic change. When the chromatin structure is dense, it will prevent the initial transcription factor and RNA polymerase from recruiting to the specific DNA sequence, which left the gene silent. However, when the chromatin structure is loose, the initial transcription factor can be recruited to the promoter region of the gene and activate the transcription of the corresponding gene. In eukaryotic cells, chromatin remodeling factors are responsible for changes in the structure of chromatin, thereby regulating gene transcription. Generally, chromatin remodeling factors are recruited to target gene sites by specific transcription factors and function as co-factors when regulating transcription initiation. For example, in mammalian cells, SWI/SNF can interact with a variety of transcription factors, such as steroid receptors, tumor suppressors and some carcinogenic factors, which can recruit chromatin remodeling complexes to the promoter region of the target gene. And then the chromatin structure of the promoter region of the target gene is changed by nucleosome sliding or replacement of the histone variant, thereby causing the activation or suppression of the corresponding gene.
In recent years, increasing attention has been paid to epigenetic regulation’s crucial role in maintaining the stability of the intracellular environment, and chromatin remodeling is an important component of it. Chromatin remodeling complexes and nucleosome dynamics are significant areas for further study. Creative BioMart provides a variety of powerful tools for epigenetic research (including high-quality recombinant proteins, synthetic peptides, nucleosomes, antibodies, cell lysates, stable cell lines, bioactive small molecules, and assay kits). We have rich experience for over 10 years and can offer various epigenetics services, such as DNA methylation analysis, RNA methylation analysis, transcriptomics profiling, histone post-translational modification analysis, chromatin remodeling and accessibility analysis, as well as epigenetic drug discovery.
References
1. Hargreaves D C, Crabtree G R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Research. 2011, 21(3): 396-420.
2. Becker P B, Workman J L. Nucleosome remodeling and epigenetics. Cold Spring Harbor Perspectives in Biology. 2013, 5(9): a017905.
3. Xu Y Z.; et al. Chromatin remodeling during host-bacterial pathogen interaction. Chromatin Remodeling. 2013.
4. Clapier C R.; et al. Mechanisms of action and regulation of ATP-dependent chromatin-remodeling complexes. Nature Reviews Molecular Cell Biology. 2017, 18(7): 407-422.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools