Creative BioMart is a leading provider of comprehensive, high quality RNA methylation analysis and is proud of offering and tailoring most types of sequencing services to meet the research objectives and budget of our clients. We hope to partner with you to explore new frontiers in biological science and accelerate your discovery.
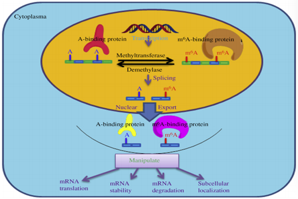
As one of the most important content of epigenetic research, RNA methylation refers to a reversible and post-transcriptional methylation modification occurring at different positions in RNA molecules and it affects several biological processes. N6-methyladenosine (m6A) and C5-methylcytidine (5mC) are two post-transcriptional modifications of RNA commonly found in eukaryotes. RNA methylation plays a key role in gene expression regulation gene expression, splicing, RNA editing, RNA stability, controlling lifespan and mRNA degradation of mRNA. Compared to DNA methylation, RNA methylation is more complex and diverse, and is ubiquitous in various eukaryotic cells. Due to the lack of effective detection methods, related studies are mostly limited to non-coding tRNA and rRNA, or a small number of coding transcripts. At the same time, most of RNA methylation functions are unknown. With the development of high-throughput sequencing technology and the discovery of some RNA methylation functions, researchers began to pay attention to RNA methylation research. In particular, the emergence of MeRIP-seq (methylated RNA immunoprecipitation sequencing) technology can efficiently and accurately detect different RNA methylation in the whole transcriptome, which lays a foundation for RNA methylation research.
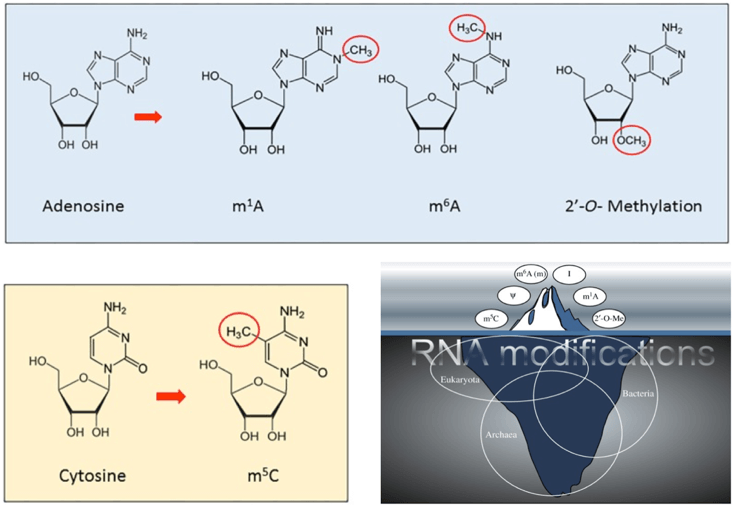
Fig 1. Several types of RNA methylation
RNA Methylation Study Procedures
To facilitate the detection of RNA methylation, Creative BioMart has established a series of methods for analyzing base methylation in RNA using bisulfite sequencing, MeRIP-seq technology and so on. Treatment of RNA with bisulfite causes the chemical deamination of nonmethylated cytosines to uracil, while methylated cytosines remain unaffected. cDNA synthesis followed by PCR amplification and DNA sequencing allows investigators to obtain the level of RNA methylation in tRNAs and rRNAs. By using high-throughput sequencing approaches, this protocol could enable the characterization of 5mC methylation patterns in RNA molecule. In addition, methylated RNA-specific antibodies are incubated with randomly interrupted RNA fragments, and methylated fragments are captured for sequencing. At the same time, a control sample needs to be sequenced in parallel to eliminate the background during the capture of methylated fragments. The two types of sequence fragments are then compared (or localized) to the reference genome/transcriptome to detect methylation level of m6A.
Our RNA Methylation Analysis Services at Creative BioMart
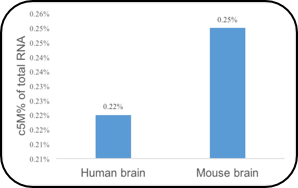
5mC RNA Sequencing
Through classical bisulfite treatment of RNA, methylation modification level of 5mC can be detected in full transcription range and in tRNA level in different species and tissues.
MeRIP-Seq Service(m6A-specific methylated RNA)
Through classical bisulfite treatment of RNA, methylation modification level of 5mC can be detected in full transcription range and in tRNA level in different species and tissues.
Global RNA 5mC Quantification
Through classical bisulfite treatment of RNA, methylation modification level of 5mC can be detected in full transcription range and in tRNA level in different species and tissues.
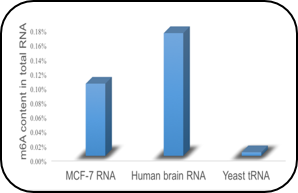
Global RNA m6A Quantification
Quantify an absolute amount of N6-methyladenosine (m6A) and determine the relative m6A RNA methylation states of RNA samples from any species such as mammals, plants, fungi, bacteria, and viruses.
With professional bioinformatics capability, Creative BioMart offers high-quality RNA methylation analysis in the whole transcriptome to identify differentially methylated regions, and ultimately help to expedite epigenetic research. If you have additional requirements or questions, please feel free to contact us.
Reference
1. Romano, G.; et al. RNA Methylation in ncRNA: Classes, Detection, and Molecular Associations. FRONT GENET. 2018, 9: 243.
2. Schaefer, M.; et al. Understanding RNA modifications: the promises and technological bottlenecks of the ‘epitranscriptome’. Open Biology. 2017, 7(5): 170077.
3. Liu, J. and Jia, G. Methylation modifications in eukaryotic messenger RNA. J GENET GENOMICS. 2014, 41: 21-33.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools