Creative BioMart provides 5mC RNA methylation sequencing service to enrich and capture methylated RNA fragments for post-transcriptional methylation studies on a whole transcriptome wide scale by classical bisulfite treatment. By identifying and analyzing the methylation site and level of samples, you can get a great deal of biological information you want.
What Is 5mC RNA Sequencing?
RNA 5mC modification is dynamically reversible. With S-adenosylmethionine (SAM) as the methyl donor, the methyltransferase can transfer the methyl group to cytosine C to form 5-methylcytosine (5mC). At present, the functional research of RNA 5mC modification is still in its infancy. However, based on the existing research, it can be speculated that 5mC is related to mRNA stability, selective splicing, retention level of exons, and assembly form of transcript. In addition, 5mC plays an important role in tRNA, including the ability to stabilize the secondary structure and enhance codon recognition of tRNA. Moreover, 5mC modification in rRNA is thought to be related to the translation process.
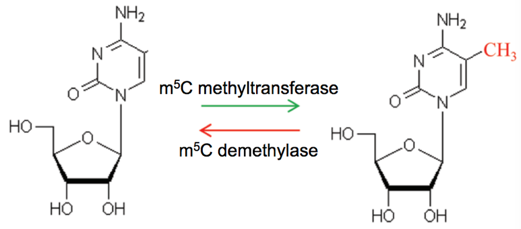
Fig 1. Dynamically reversible process of 5mC RNA methylation
The 5mC modification at the overall level of RNA can be detected by mass spectrometry, and there are four methods for the identification of 5mC single point, which are 5mC bisulfite sequencing (BS-seq), 5mC-RIP, Aza-IP, and miCLIP. Among them, RNA bisulfite sequencing is the most ideal method so far. Bisulfite treatment can remove the amino group of cytosine C (which accounts for the majority) that is not methylated in the nucleotide sequence and converts it into uracil U, while methylated cytosine C remains unchanged. After PCR amplification, uracil U is completely converted to thymine T, and thus it can be distinguished from the cytosine C by original methylation modification. Finally, the PCR product is sequenced and compared to the untreated sequence to determine if the cytosine site has been methylated.
5mC RNA Sequencing Workflow
The general workflow for 5mC RNA sequencing is outlined below. Briefly, it consists of six steps:
(I) RNA sample fragmentation
(II) Bisulfite treatment of fragmented RNA samples
(III) Reverse transcription of bisulfite-treated RNA samples to synthesize cDNA
(IV) Construction of sequencing library with cDNA
(V) High-throughput sequencing
(VI) Data analysis of sequencing results to obtain 5mC methylation information of RNA
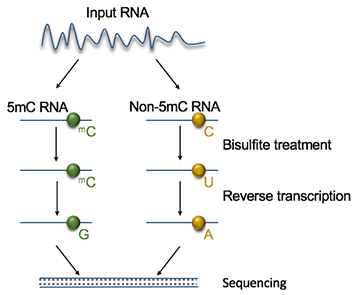
Fig 2. Diagram of 5mC RNA Sequencing workflow
Categories of 5mC RNA Sequencing
Advantages of Creative BioMart
With professional bioinformatics capability, Creative BioMart’ high-quality 5mC RNA sequencing services are tailored to each project to ensure the objectives to be met and help to expedite epigenetic research. Our expert project teams are assigned to each study with a focus on completing projects in the shortest amount of time.
If you have additional requirements or questions, please feel free to contact us.
Reference
1. Romano, G.; et al. RNA Methylation in ncRNA: Classes, Detection, and Molecular Associations. FRONT GENET. 2018, 9: 243.
2. Nachtergaele, S. and He, C. The emerging biology of RNA post-transcriptional modifications. RNA Biology. 2017, 14(2): 156–163.
3. Yang, X.; et al. 5-methylcytosine promotes mRNA export – NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Research. 2017, 27(5): 606-625.
4. Liu, J. and Jia, G. Methylation Modifications in Eukaryotic Messenger RNA. J GENET GENOMICS. 2014, 41(1), 21–33.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools