As a leading provider of epigenetic analysis services, Creative BioMart is able to offer MRE-Seq service, which is based on methylation-sensitive restriction endonuclease digestion and next-generation sequencing, to help customers mine the information on the unmethylated CpG loci from MRE-Seq high-throughput data.
What Is MRE-Seq?
MRE-Seq represents methylation-sensitive restriction enzyme digestion followed by sequencing. First, several methylation-sensitive restriction endonucleases are required to digest intact genomic DNA. These methylation-sensitive restriction enzymes (MREs), such as BstUI, HpaII, NotI and SmaI, only cleave the unmethylated CpG sites of genomic DNA and keep the methylated sites intact. The obtained DNA fragments are then subjected to size selection and large-scale parallel sequencing. The sequencing of the digested DNA fragments enriched with unmethylated CpGs at their ends allows identification of the cutting sites, and when mapped back to the reference genomic assembly, these sequencing readings reveal the location of the unmethylated CpGs within the recognition sites of the enzymes utilized. MRE-Seq can cover approximately 30% of the genome by utilizing multiple cut-sites. Although MRE-Seq allows for estimation of relative DNA methylation levels, there is relatively low genomic coverage because the recognition sites containing CpGs are limited. Furthermore, while methylated CpGs can be inferred by lack of readings at the cleavage sites, it still needs to be processed in cases where complete digestion is guaranteed.
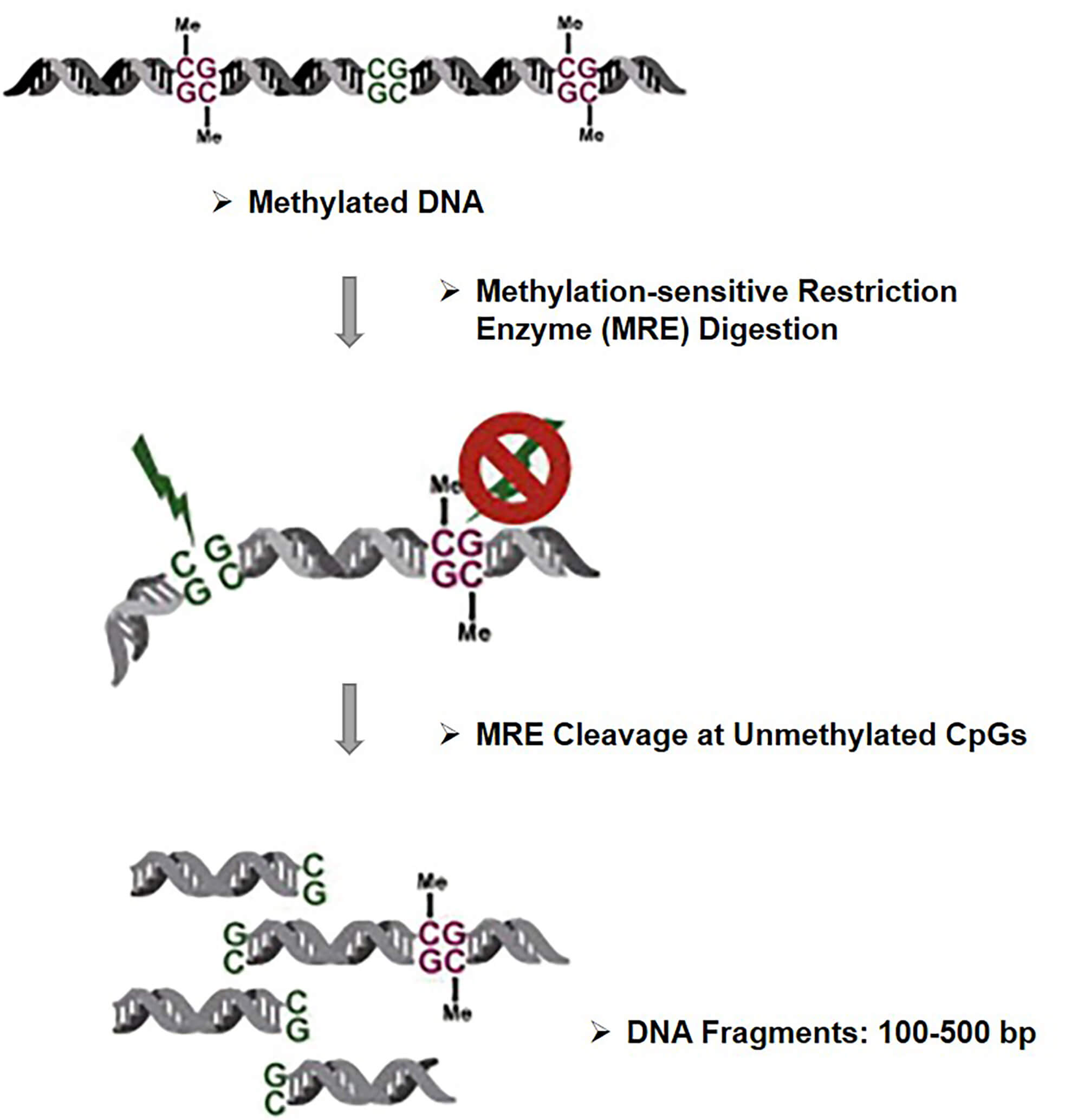
Fig 1. The principle of MRE
Features of MRE-Seq
Our Advantages
Workflow of MRE-Seq at Creative BioMart
In the workflow of MRE-Seq, the MREs cleave the unmethylated CpG sites of genomic DNA, and the resulting DNA fragments are size-selected and sequenced. During the process of MRE-Seq, we strictly control every step to ensure access to reliable experimental data.

Fig 2. Workflow of MRE-Seq at Creative BioMart
Creative BioMart not only provides independent MRE-Seq service, but also identifies different methylation regions (DMRs) by jointly modeling MRE-Seq and MeDIP-Seq data. Both MRE-Seq and MeDIP-Seq are techniques for measuring DNA methylation at the genome-wide level. They represent complementary manner of enriching the unmethylated portion of the genome or the methylated portion of the genome. Both techniques have their own pros and cons and can be applied individually or in combination. MRE-Seq provides DNA methylation estimates at single CpG resolution but is considered low coverage due to the limit of CpG containing recognition sites. An important advantage of MeDIP over enzymatic digestion-based methods is the lack of bias for specific nucleotide sequence, other than CpG, but its resolution is limited by the size of the immunoprecipitated DNA fragments. Because MRE-Seq and MeDIP-Seq are independent and complementary, combining these two techniques can yield synergistic advantages. At Creative BioMart, our experts utilize methylCRF, a conditional random fields-based algorithm, to integrate MRE-Seq and MeDIP-Seq data to predict genome-wide DNA methylation levels at single CpG resolution.
With experienced experts and fast-responding teams, Creative BioMart can provide high-quality MRE-Seq service, and the most reliable and comprehensive data is offered timely to the customers during the experiments. If you have additional requirements or questions, please don’t hesitate to contact us.
References
1. Maunakea A K.; et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010, 466(7303): 253-257.
2. Li D.; et al. Combining MeDIP-seq and MRE-seq to investigate genome-wide CpG methylation. Methods. 2015, 72: 29-40.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools