Creative BioMart is able to offer MeRIP-Seq (m6A-specific methylated RNA) service to help customers exploit RNA methylation patterns from MeRIP-seq high-throughput data, reveal the potential functions of mRNA methylation in regulating gene expression, splicing, and translation, and effectively guide an amount of basic research and the intervention of various diseases.
What Is m6A Methylation and MeRIP-Seq?
N6-methyladenosine (m6A) is the most common form of RNA methylation, accounting for 80%. m6A is dynamically reversible and has important research significance, since it plays a key role in post-transcriptional regulation, including regulation of gene expression, splicing, RNA editing, RNA stability, control of mRNA lifetime and degradation, and mediation of circular RNA translation. Therefore, in-depth study of m6A RNA methylation can help solve biological problems such as cell differentiation, biological growth and development, and disease development, and help drug developers design small molecules that regulate gene expression, and kill or control diseased cells. Methylated RNA immunoprecipitation sequening (MeRIP-seq) technology can be used to comprehensively study the post-transcriptional methylation modification of RNA, which is a key technology for epigenetic research.
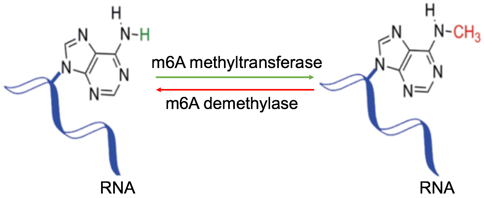
Fig 1. Dynamically reversible process of m6A RNA methylation
MeRIP-seq technology combines methylated DNA immunoprecipitation (MeDIP) technology, RNA immunoprecipitation (RIP) technology and RNA sequencing (RNA-seq) technology to accurately detect RNA methylation in the whole transcriptome range. The methylated RNA-specific antibodies are incubated with randomly interrupted RNA fragments by immunoprecipitation, and the methylated fragments are captured for sequencing. At the same time, a control sample needs to be sequenced in parallel, and is used to eliminate the background in the process of capturing the methylated fragment. The sequence fragments in the immunoprecipitation (IP) sample and the control sample are then compared or localized to the reference genome/transcriptome to detect the RNA methylation site. The control sample measures the expression quantity of the corresponding RNA, which is essentially RNA-seq data.
MeRIP-Seq Workflow
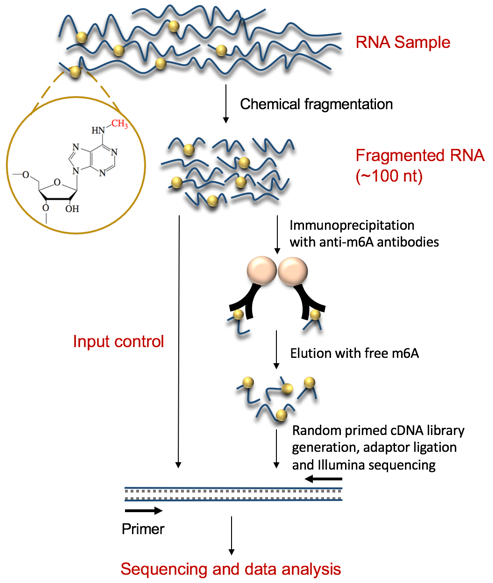
Fig 2. Diagram of MeRIP-Seq workflow
Categories of MeRIP-Seq
Features of MeRIP-Seq
Advantages of Creative BioMart
With experienced experts and fast-responding teams, Creative BioMart can provide high-quality MeRIP-Seq services, including sample standardization, library construction, deep sequencing, raw data quality control, and customized bioinformatics analysis, to ensure the objectives are met and expedite epigenetic research. If you have additional requirements or questions, please feel free to contact us.
Reference
1. Wang, X.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 2014, 505(7481):117-20.
2. Liu, J. and Jia, G. Methylation Modifications in Eukaryotic Messenger RNA. J GENET GENOMICS, 2014, 41(1): 21-33.
3. Cao, G.; et al. Recent advances in dynamic m6A RNA modification. Open biology, 2016, 6(4): 160003.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools