95% of the human genome does not actually encode proteins but produces a large number of non-coding RNAs that play an important regulatory role in the growth and development of life and are closely related to various diseases such as HIV/AIDS, leukemia, diabetes and deformity, as well as to participate in the regulation of stem cells and epigenetics. RNA-protein complexes drive post-transcriptional regulation of gene expression in almost all cellular processes, including splicing, nuclear export, mRNA stability, and protein translation. Therefore, the understanding towards post-transcriptional regulatory networks and mechanisms depends on determining the changes in the binding of RNA in these processes.
In addition, RNA-binding proteins control many aspects of cell biology by binding to single-stranded RNA binding motifs (RBMs). However, RBMs can be buried within their local RNA structure, thereby inhibiting RNA-protein interactions. Through the analysis of RNA-protein interactions, the ‘m6A-switch” mechanism has been discovered. This mechanism involves N6-methyladenosine (m6A), the most abundant and dynamic internal modification of eukaryotic messenger RNA (mRNA) alter local structure of the mRNA and the long non-coding RNA (lncRNA), promoting the binding of heterogeneous nuclear ribonucleoprotein C (HNRNPC), a rich nuclear RNA-binding protein that is responsible for pre-mRNA processing, and thus affecting the regulation of gene expression and RNA maturation. At present, the study of the interaction between RNA and protein has been paid more and more attention by scientists and has become a hot field in epigenetic research.
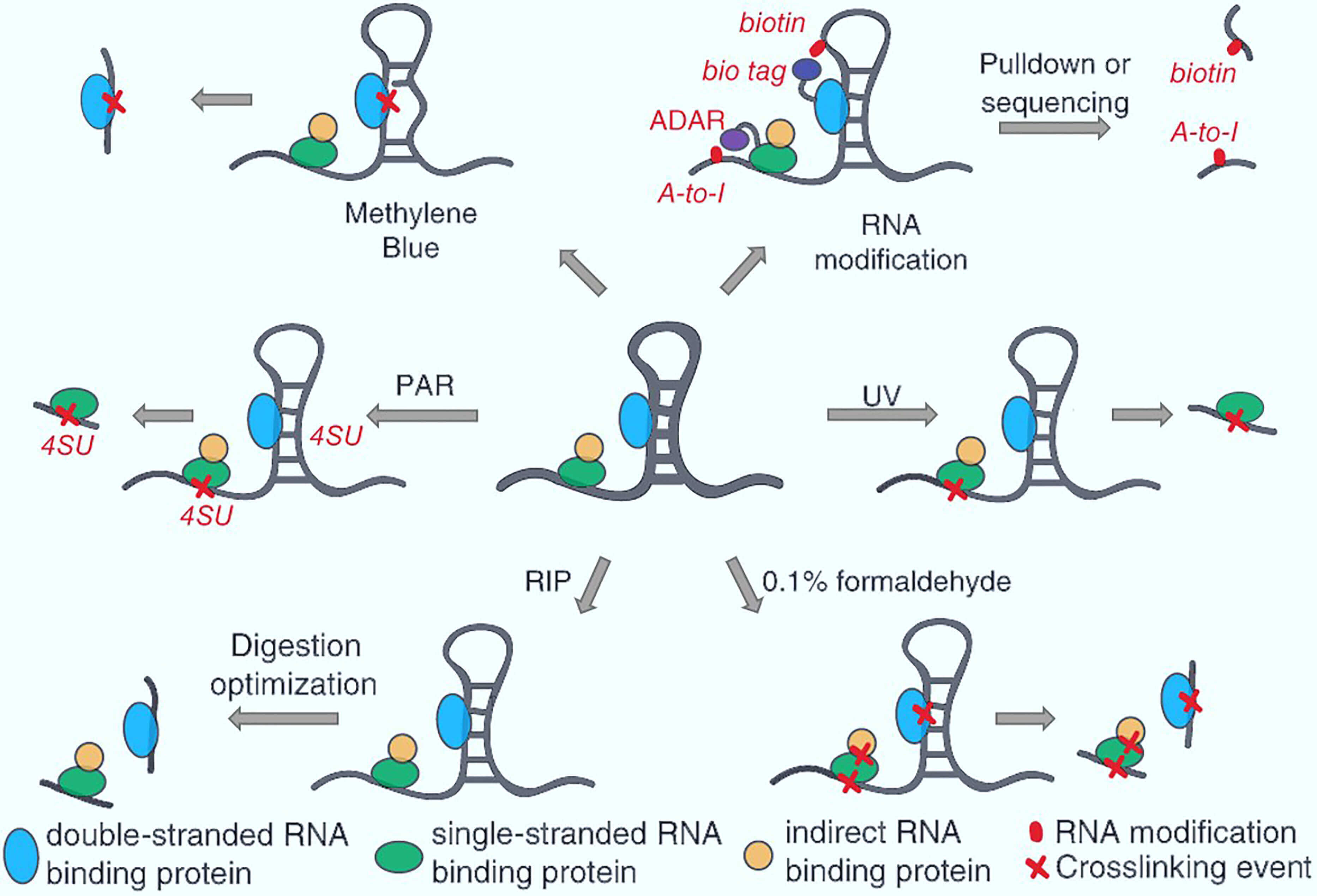
Figure 1. Schematic illustration of RNA-protein interaction analysis methods
Creative BioMart has organized a team of experts with extensive experience in RNA-protein interaction analysis to provide flexible and one-stop services for your RNA-protein interaction project. We are capable of helping you with your project consultation, experiment design, carry out your experiments, and perform data analysis. A series of detection methods including RNA immunoprecipitation (RIP) and crosslinking-immunoprecipitation (CLIP) have been established in our lab to help with your research. You can choose the technology you prefer, and we can also help recommend the most appropriate solution. Moreover, integrated services are also offered to obtain the most comprehensive and in-depth research results.
RNA-protein Interaction Analysis Services at Creative BioMart
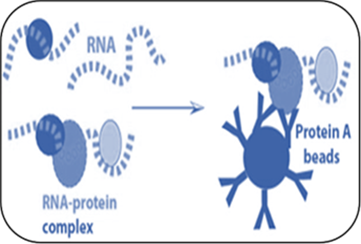
RNA Immunoprecipitation (RIP) Service
RIP precipitates a specific RNA-binding protein (RBP) and associated RNAs which can be analyzed by downstream applications, such as real-time PCR, microarrays or next-generation sequencing. Creative BioMart offers RIP service together with microarray (RIP-chip/rRIP-chip) and sequencing (RIP-Seq) to identify RNAs bound by the RBP of interest.
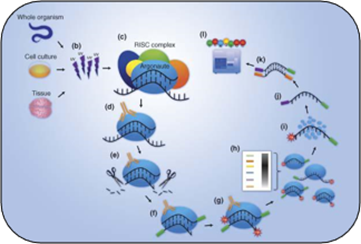
Crosslinking-immunoprecipitation (CLIP) Service
CLIP is performed by immunoprecipitation with an antibody against the protein of interest after UV cross-linking of the RNA-protein complex. At Creative BioMart, various CLIP techniques are provided, including photoactivatable ribonucleotide-enhanced CLIP (PAR-CLIP), individual nucleotide-resolution CLIP (iCLIP/miCLIP), HITS-CLIP (also known as CLIP-Seq).
With experienced technical team and state-of-art technology platform, Creative BioMart has the confidence to work with you to study RNA-protein interaction at a flexible and one-stop manner, including program consultation, experiment design, experimental operation, data analysis, as well as trouble-shooting. In addition, a customized combination of services is provided for comprehensive and in-depth RNA-protein interaction analysis.
Creative BioMart offers our RNA-protein interaction analysis services to many customers worldwide in the biopharmaceutical and biotech industries. We are eager to learn of your specific requirements and to speed up your academic research or product development. Please don’t hesitate to contact us if you have any questions regarding our service.
Reference
1. Liu N.; et al. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015, 518(7540): 560-564.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools