If you don’t want to chop up your DNA samples with enzymes, or treat them with bisulfite, which can damage DNA and lead to inaccuracies in downstream applications, Creative BioMart offers you an enrichment-based approach. Methylated DNA immunoprecipitation (MeDIP) is an antibody-based, large-scale technique used to enrich and capture methylated DNA fragments for gene-specific DNA methylation studies on a genome wide scale. Following the MeDIP, DNA methylation can be analyzed using multiple downstream applications, including MeDIP-Seq, MeDIP-PCR and MeDIP-chip.
What Is MeDIP?
Methylated DNA immunoprecipitation (MeDIP) is a technique that utilize 5mC-specific antibody to capture and enrich methylated DNA. In other words, the fragmented genomic DNA is incubated and precipitated together with the 5mC-specific antibodies. The methylated DNA fragments after enrichment can be used for microarray or sequencing, etc. This technique is able to detect highly methylated regions in the genome but cannot perform DNA methylation analysis at single-resolution. Like another approach based on affinity enrichment, the methyl-CpG binding domain (MBD) protein capture method, both have significant advantages over bisulfite-based techniques. First, 5mC and 5hmC can be distinguished. Moreover, DNA will not be damaged as much as bisulfite treatment. Comparing MeDIP and MBD protein capture, MeDIP is more sensitive to methylation differences in areas with low CpG density, while MBD is more sensitive in areas with higher CpG density, such as CpG islands.
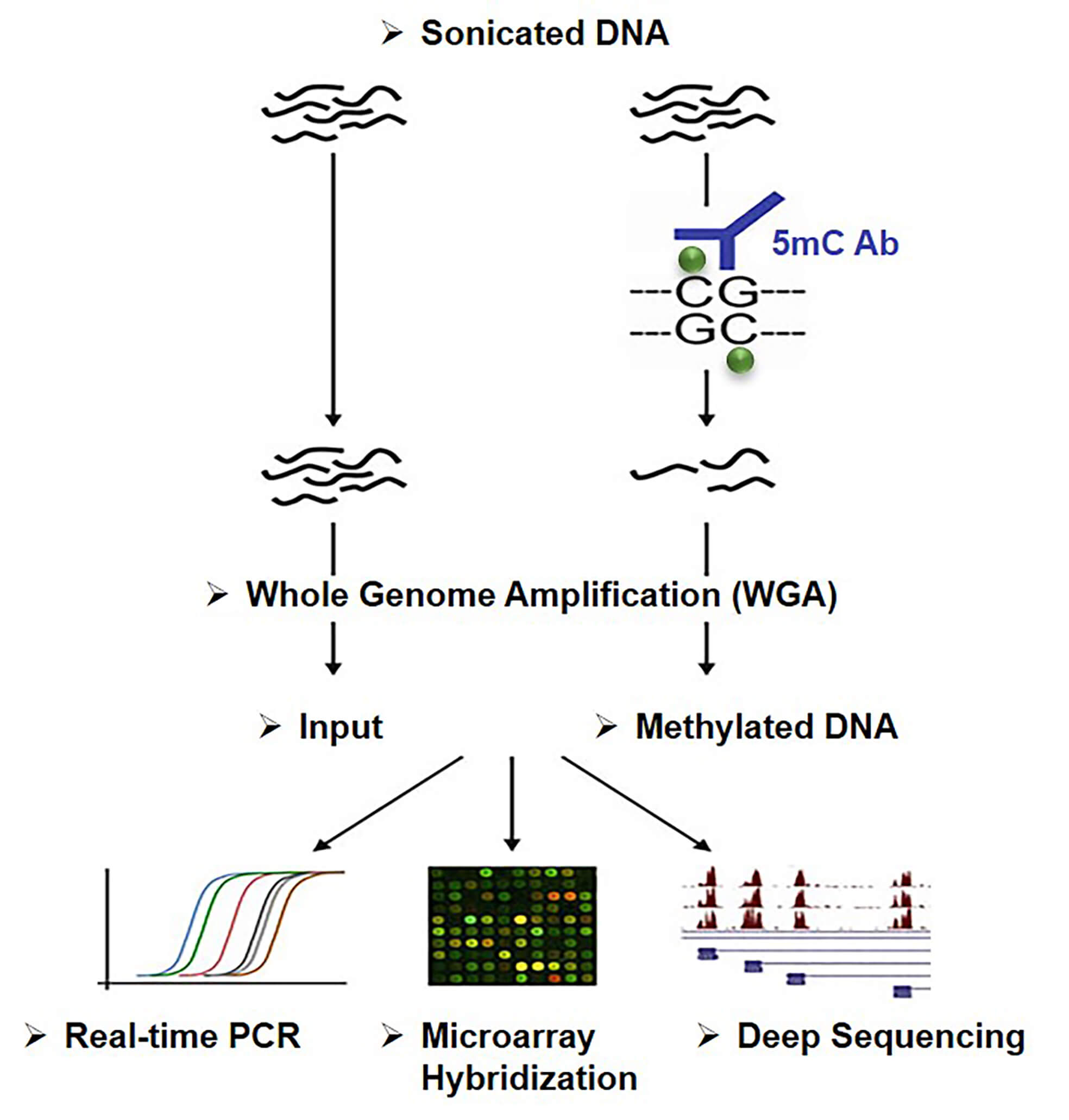
Figure 1. Principle of MeDIP
Features of MeDIP
MeDIP-based Services at Creative BioMart
MeDIP-Seq Service
MeDIP-Seq stands for methylation-dependent immunoprecipitation followed by sequencing. MeDIP-Seq produces relative enrichment of methylated DNA across the entire genome, rather than predicting absolute DNA methylation levels. Deep sequencing provides greater genomic coverage and represents the majority of immunoprecipitated methylated DNA.
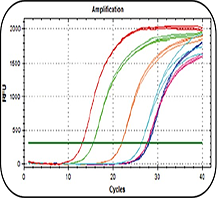
MeDIP-qPCR Service
The methylated DNA fragments are enriched through MeDIP approach, and then detected by real-time quantitative PCR. The accurate value of methylation level of the specific site in the detected biological sample is obtained through data analysis and processing. This technique allows primers to be designed for gene promoters or CpG islands of interest.

MeDIP-chip Service
The methylated DNA immunoprecipitation microarray (MeDIP-chip) is a genome-wide, high-resolution approach for detecting DNA methylation in whole genome or CpG islands. The MeDIP enriched methylated DNA can be interrogated using DNA microarrays. This combined approach allows researchers to rapidly identify methylated regions in a genome-wide manner.
With a skillful technical team, Creative BioMart can provide high-quality MeDIP-based services, ensuring the objectives are met and accelerate your epigenetic research. We provide a complete report with both raw and analyzed data, graphs, detailed protocols, and a summary of findings. contact us for more information about our MeDIP-based services.
References
1. Borgel J.; et al. Methylated DNA immunoprecipitation (MeDIP) from low amounts of cells. Methods in Molecular Biology. 2012, 925: 149.
2. Nair S S.; et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011, 6(1): 34-44.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools