Our custom Global RNA m6A Quantification service provides personalized solutions designed to supply researchers with two types of detection methods to meet their specific needs with the advantages of high quality, efficiency and cost-saving. We can help you decide on the most suitable quantification service or approach for your applications and optimize detecting procedures toward your specific need. We will find a solution for you at Creative BioMart.
Introduction of Global RNA m6A Quantification
In eukaryotes, 5′-end Cap and 3′-ployA modification play an important role in transcriptional regulation. Currently, known types of enzymes involved in the modification of N6-methyladenosine (m6A) that are already known include demethylases, methylases, and methylation recognition enzymes. An abnormality in the enzyme involved in m6A modification will cause abnormal methylation in RNA, which leads to a series of diseases including obesity, tumor, neurological disease, and embryonic developmental delay. In addition, some non-coding RNAs such as lncRNA, tRNA, rRNA and splicing RNA also have a large number of base modification activities before and after transcription. These physiological processes can regulate gene expression and mediate RNA translation. Because the levels of m6A are changed in eukaryotic cells of different types and different states, quantitative detection of m6A modification in the transcriptome can provide researchers with a large amount of bioinformatics data, which is also beneficial for the diagnosis and treatment of diseases.
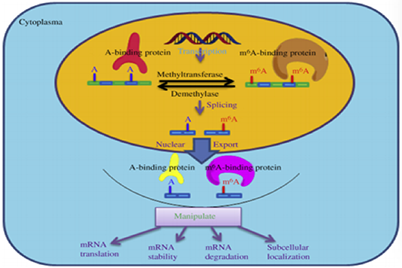
m6A RNA methylation modification can be quantitatively detected by both colorimetric and fluorescent methods, which is suitable for direct detection of total RNA methylation status isolated from any species, such as mammals, plants, fungi, bacteria and viruses. In this assay, RNA samples are incubated with the antibodies that can specifically capture N6-methyladenosine. The absorbance is read at a wavelength of 450 nm in a spectrophotometer while the detection signal is enhanced and then quantified by colorimetry. The amount of m6A is proportional to the measured OD intensity. Alternatively, the RFU (relative fluorescence unit) is read in a fluorescence spectrophotometer for fluorescence measurement. And the amount of m6A is also proportional to the measured fluorescence intensity.
Global RNA m6A Quantification Workflow
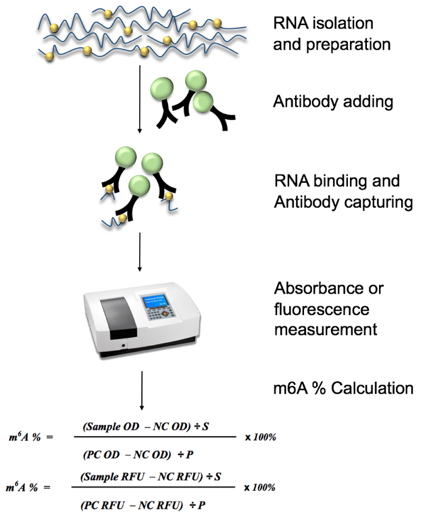
Fig 1. Diagram of Global RNA m6A Quantification workflow
Advantages of Creative BioMart
Supported by our experienced experts, fast-responding teams and advanced technology, Creative BioMart can help you to acquire global m6A RNA methylation information at one time by strict quality control and advanced bioinformatics analyses. Please contact us for more information or a detailed quote.
Reference
1. Wang, X.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014, 505(7481):117-20.
2. Fu, Y.; et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014, 15(5): 293-306.
3. Alarcón, C. R.; et al. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015, 519(7544): 482-5.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools