The structure and dynamics of nucleosomes clearly participate in the regulation of DNA transactions such as transcription and replication. Histone and/or DNA modifications can modulate the structure and dynamics of nucleosomes and consequently alter the transcriptional outcome. These events can be thought of as intrinsic dynamics. Revealing the structural dynamics of nucleosomes is a key step in elucidating the mechanisms that regulate genomic accessibility.
Before the advent of single-molecule technology, kinetic experiments could only represent the average behavior of a large number of molecules in the measurement time, while the organism is a complex, dynamic and heterogeneous system. Therefore, monitoring the dynamics of single molecules is of great significance. Single-molecule fluorescence resonance energy transfer (smFRET) technology studies the conformational changes of molecules by detecting the efficiency of fluorescence energy transfer between donor and acceptor within a single molecule. The multicolor stochastic optical reconstruction microscopy is implemented using smFRET, monitoring nucleosomes in real-time at the nanometer level to obtain relevant kinetic data.
Creative BioMart currently has the ability to provide smFRET technology to support the study on the structure and structural dynamics of nucleosomes.
What Is smFRET?
FRET is an energy transfer phenomenon that occurs between two fluorescent molecules that are relatively close to each other. When the emission spectrum of the donor overlaps with the absorption spectrum of the acceptor, and the two molecules are within 10 nm apart, a non-radioactive energy transfer will occur, weakening the fluorescence intensity of the donor, while enhancing the fluorescence emitted by the receptor. The FRET efficiency is closely related to the spatial distance of the donor and acceptor molecules. And as the distance is extended, the FRET efficiency is significantly decreased.
FRET between a pair of single fluorophore molecules. The FRET distance within a single molecule can be monitored by recording the fluorescence intensity of a single FRET pair labeled at a single molecule or complex. If the molecule of interest is immobilized on a surface, a FRET time trajectory of the single molecule can be constructed, which represents the time-resolved dynamics of the molecule.
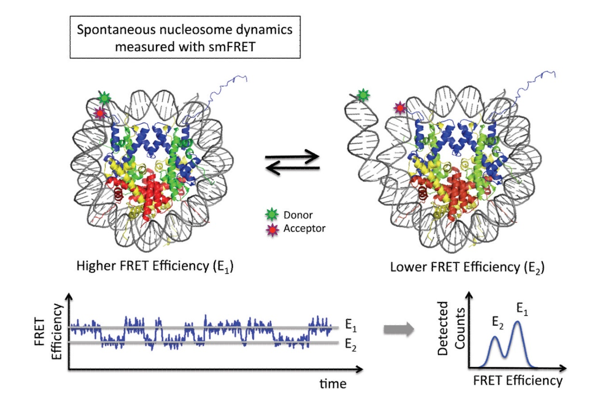
Figure 1. smFRET Studies on the Structure and Structural Dynamics of Nucleosomes. (Choy J S.; et al. 2012). The time-averaged FRET efficiency histogram reveals the closed (E1; high FRET) and opened (E2; low FRET) states of a nucleosome.
Features of smFRET
Our Advantages
Workflow of smFRET Service at Creative BioMart
Figure 2. Workflow of smFRET Service at Creative BioMart
If you have any special needs in studying the structure and structural dynamics of nucleosomes by smFRET, please contact us for this special service. Let us know what you need, and we will meet your requirements. We look forward to working with you.
Reference
1. Choy J S, Lee T H. Structural dynamics of nucleosomes at single-molecule resolution. Trends in Biochemical Sciences. 2012, 37(10): 425-435.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools