Brief Introduction
DNA methylation is the most basic epigenetic modification. In eukaryotes, the most common methylation modification is 5-methylcytosine (5mC), which is generated by attaching the methyl group to the fifth carbon of the pyrimidine ring of CpG island cytosine with the help of DNA methyltransferase (DNMT). Besides, DNA methylation occurs at the N-6 position of adenine (N6-methyladenine, 6mA), the N-4 position of cytosine (N4-methylcytosine, 4mC), as well as the N-7 position of guanine (N7-methylguanine, 7mG), which are respectively catalyzed by different DNA methyltransferases. The gene sequence is not altered during this process, but it regulates gene expression. DNA methylation is a dynamic process, and the DNA demethylation is mediated by DNA demethylases.
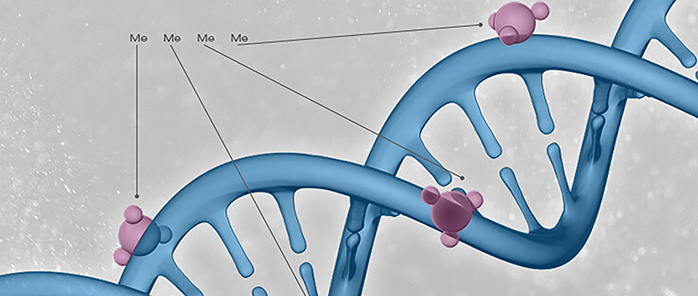
Considerable literature has shown that DNA methylation affects the development of human organs, genetic diseases, and the development and progression of tumors. The traditional view is that the CpG island methylation plays a role in transcriptional repression, because transcription factors (TFs) often bind to DNA motifs without methylation, and methylation can directly interrupt the combination of the two. Alternatively, the methylated regions of DNA (in a sequence-dependent manner) are competitively bound by MDBPs (methyl-CpG binding-domain proteins).
In addition to DNA methylation, some studies have identified variants of DNA methylation, such as DNA hydroxymethylation mediated by TET (ten eleven translocation).
Creative BioMart provides a variety of DNA methyltransferases, DNA demethylases, and methylated DNA-binding proteins, which are involved in DNA methylation, DNA hydroxymethylation, and other DNA modifications.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools