Chromatin immunoprecipitation (ChIP) has been routinely used to investigate changes in transcription factor binding and histone modifications, which are expected to correlate with changes in gene expression. To determine the biological significance of changes detected by ChIP, gene expression studies are always performed in parallel. The comparison of RNA expression microarray data with ChIP data is a common approach to study these changes, while it is not ideal since the data is not derived from the exact same sample. Moreover, RNA microarray data can not provide information about alternative transcription start sites, making the association of transcription factor binding sites with specific genes error prone. Machanisms of post-transcriptional regulation also have impact on RNA in manners that may not accurately reflect the transcription changes. In view of these problems, Creative BioMart provides an alternate method, RNA polymerase II (Pol II) ChIP-Seq to RNA-based methods that measure gene expression.
The transcription rates across the genome can be measured in a single Pol II ChIP-Seq experiment since occupancy of Pol II is strongly associated with gene transcription rates. RNA Pol II ChIP-Seq is a valuable supplement to any transcription factor or histone ChIP experiment, allowing researchers to correlate binding of transcription factors or histones with gene transcription. Unlike other RNA-based techniques, Pol II ChIP-Seq service at Creative BioMart enables you to measure transcription rates without the influence of RNA half-life. It has additional advantages of using the same material that is utilized in your transcription factor targeted ChIP experiments. Therefore, it helps you to combine transcription factors binding with transcriptional changes in the same assay.
Features of Pol II ChIP-Seq Service
Workflow of Pol II ChIP-Seq Service at Creative BioMart
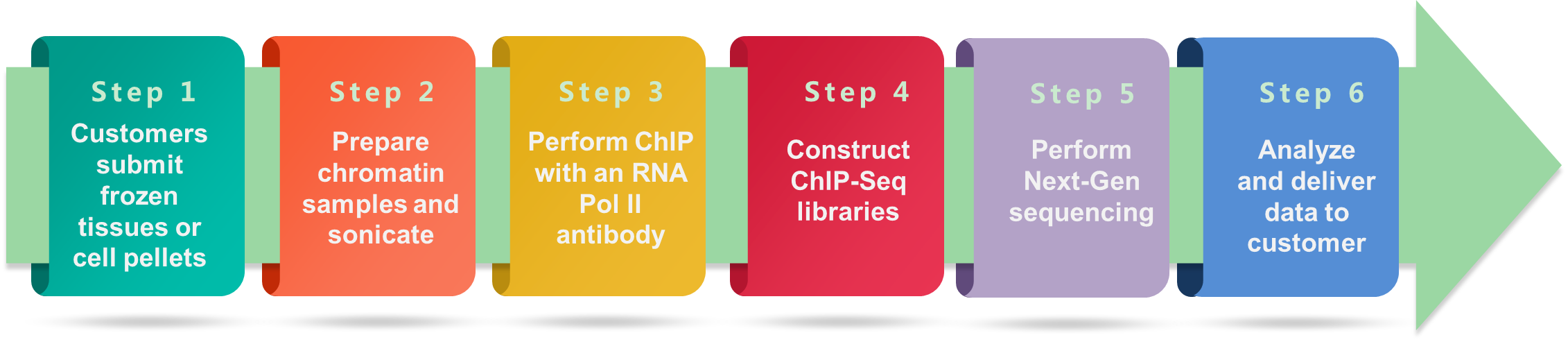
Figure 1. Schematic workflow of Pol II ChIP-Seq service
Creative BioMart is committed to becoming a leader in the field of epigenetics and to making ChIP-Seq service available to scientists in academic labs and biopharmaceutical industries. Equipped with state-of-art technology platforms and skilled experts, Creative BioMart could overcome the technical and bioinformatics challenges in generating whole-genome data sets as well as provide our customers high-quality and interpretable data from our Pol II ChIP-Seq. contact us now and tell us what your demand is!
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools