If your research goal is to reveal and map the genome-wide distribution of newly discovered epigenetic markers 5fC and 5caC that may play a key role in epigenetics, or to quantify TDG-mediated excision of 5fC/5caC at high resolution, Creative BioMart currently offers methylation-assisted bisulfite sequencing (MAB-Seq) technology that allows base-resolution mapping of 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), two oxidized 5-methylcytosine (5mC) bases indicative of active, and measurement of their relative abundance.
What Is MAB-Seq/caMAB-Seq?
In MAB-Seq, genomic DNA is first treated with the bacterial DNA CpG methyltransferase M.SssI that efficiently methylates cytosines (C) within CpG dinucleotides. Bisulfite conversion of M.SssI-treated DNA only deaminates 5fC and 5caC; the originally unmodified C within CpGs becomes resistant to bisulfite conversion due to its conversion to 5mC. In subsequent sequencing, only 5fC and 5caC are read as T, while C/5mC/5hmC are read as C. Based on MAB-Seq, the caMAB-Seq (5caC methylase-assisted bisulfite sequencing) is further developed to directly map 5caC at single-base resolution. In this modified version of MAB-Seq, genomic DNA is first treated with sodium borohydride (NaBH4) to reduce 5fC back to 5hmC. Owing to the combination of NaBH4 reduction with M.SssI treatment, 5caC is directly mapped as T after bisulfite conversion, while C/5mC/5hmC/5fC are read as C.
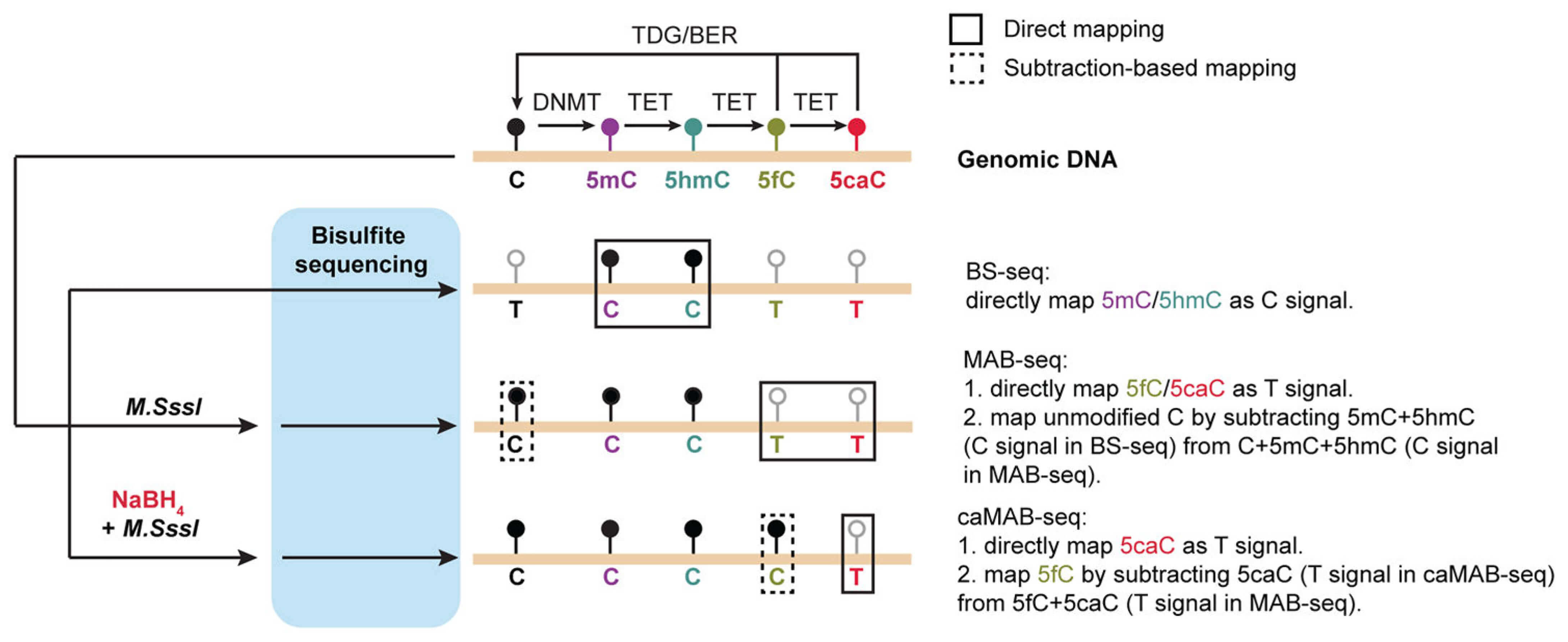
Figure 1. Schematic Illustration of the Principle behind MAB-Seq/caMAB-Seq. (Wu H.; et al. 2016).
Features of MAB-Seq
Advantages of Choosing Creative BioMart
Workflow of Our Service at Creative BioMart

Figure 2. Workflow of Genome-wide Profiling of 5fC and 5caC at Creative BioMart
With skillful experts and stringent quality control, genome-wide profiling of 5fC and 5caC service provided by Creative BioMart is ready for any customer demands with optimal customization. If you have any questions about this service, please don’t hesitate to contact us.
References
1. Neri F.; et al. Methylation-assisted bisulfite sequencing to simultaneously map 5fC and 5caC on a genome-wide scale for DNA demethylation analysis. Nature Protocols. 2016, 11(7): 1191-1205.
2. Wu H.; et al. Base-resolution profiling of active DNA demethylation using MAB-seq and caMAB-seq. Nature protocols. 2016, 11(6): 1081-1100.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools