Genome-wide analysis helps understand the functional effects of DNA methylation on phenotypic plasticity, which in turn requires a technique of balancing accuracy, genome coverage, resolution and cost, but low DNA input to minimize the consumption of precious samples. Methylation DNA immunoprecipitation sequencing (MeDIP-Seq) provided by Creative BioMart meets these criteria, combining MeDIP with massively parallel DNA sequencing.
What Is MeDIP-Seq?
MeDIP-Seq represents methylation-dependent immunoprecipitation followed by sequencing. MeDIP-Seq produces relative enrichment of methylated DNA across the entire genome, rather than predicting absolute DNA methylation levels. In a typical MeDIP-Seq protocol, monoclonal antibodies directed against 5mC are used to enrich size-selected methylated DNA fragments (usually 100-300 bp), which are then sequenced and analyzed. Deep sequencing provides greater genomic coverage and represents the majority of immunoprecipitated methylated DNA. Antibody-based selection does not enrich 5hmC due to antibody specificity. Like methyl-CpG-binding domain sequencing (MBD-Seq), MeDIP-Seq is also able to detect differentially methylated regions (DMRs) and capture approximately the same proportion of methylome. However, proteins with the methylated CpG binding domain (MBD) bind strictly to methylated CpG, while antibodies used in MeDIP capture DNA fragments contain any methylated cytosines, where non-CpG methylation may be important for some diseases. One of the inherent limitations of MeDIP-Seq is that the resolution is affected by the size of the immunoprecipitated methylated DNA fragments, but since the methylation status of adjacent CpGs is correlated, MeDIP-Seq is a cost-effective approach when single-base resolution is not required. Moreover, MeDIP-Seq is feasible even with a small amount of input DNA.
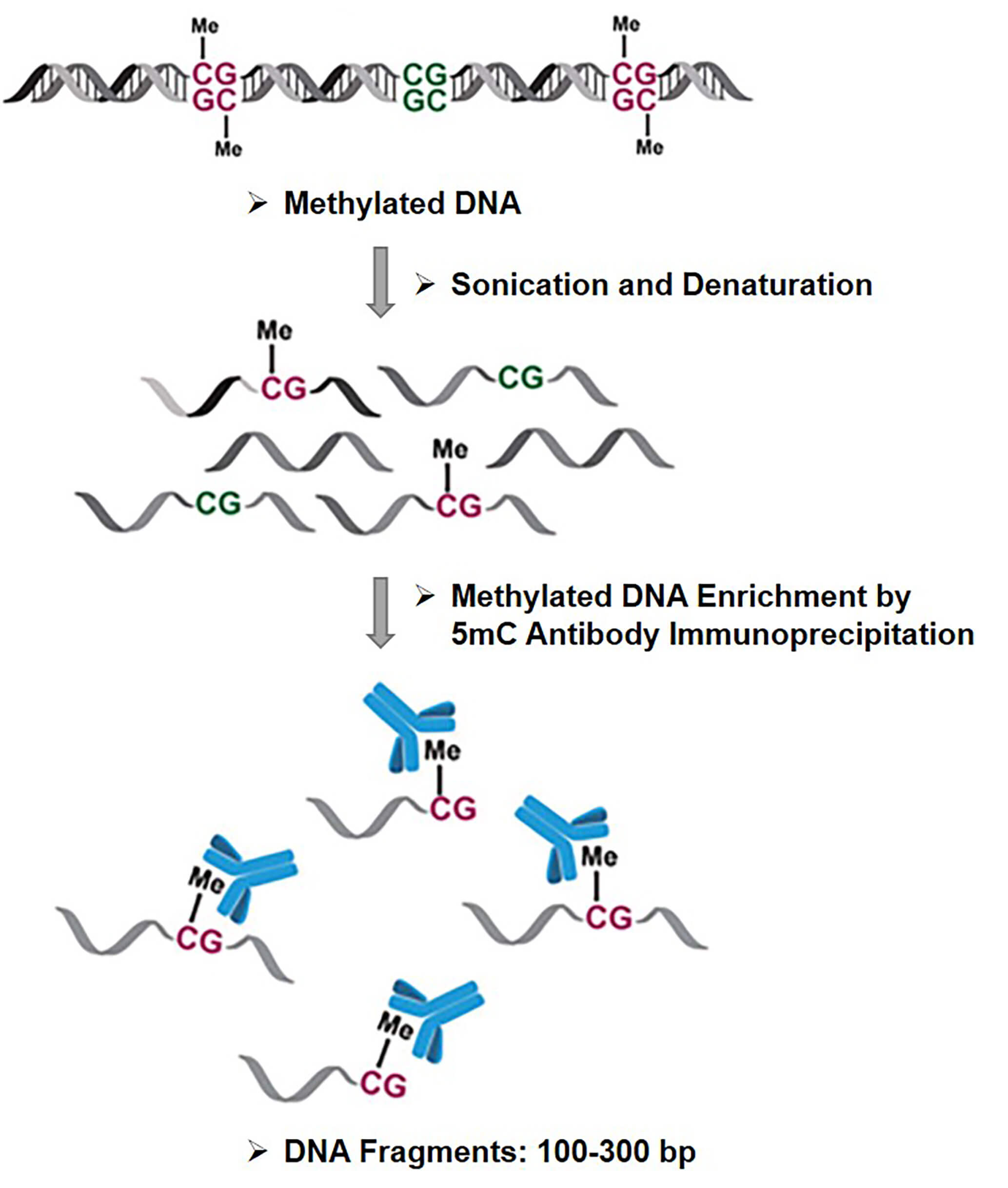
Fig 1. The schematic diagram of MeDIP
Features of MeDIP-Seq
Our Advantages
Workflow of MeDIP-Seq at Creative BioMart
Briefly, the extracted DNA is fragmented, denatured, ligated with adaptor and captured using the 5mC antibody. The enriched methylated DNA fragments are then amplified and sequenced. Through optimized experimental procedures and our highly specific 5mC antibodies for enriching methylated DNA fragments, you can complete genome-wide DNA methylation studies with extremely high efficiency and specificity at a lower cost.

Fig 2. Workflow of MeDIP-Seq at Creative BioMart
Additionally, at Creative BioMart, our experts can utilize computational algorithms (i.e., M&M and methylCRF) to integrate data from the complementary techniques of MRE-Seq and MeDIP-Seq to compensate for the inherent limitations of MeDIP-Seq, thus predicting genome-wide DNA methylation levels at single CpG resolution.
With experienced experts and first-class technical platform, Creative BioMart can provide high-quality MeDIP-Seq service, ensuring to accelerate your epigenetic research. If you have additional requirements or questions, please feel free to contact us.
References
1. Taiwo O.; et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nature Protocols. 2012, 7(4): 617-636.
2. Nair S S.; et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011, 6(1): 34-44.
3. Li D.; et al. Combining MeDIP-seq and MRE-seq to investigate genome-wide CpG methylation. Methods. 2015, 72: 29-40.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools