Fully understanding the potential function of 5-hydroxymethylcytosine (5hmC, a modification of DNA cytosine in mammalian cells) requires a precise single-base resolution sequencing method. Creative BioMart provides Tet-assisted bisulfite sequencing (TAB-Seq), which is based on bisulfite sequencing and identifies 5hmC at a single-base resolution and determines its abundance at each modified site.
What Is Tet-Assisted Bisulfite Sequencing?
Bisulfite sequencing is recognized as the “gold standard” for DNA methylation sequencing. Treatment of DNA with sodium bisulfite converts unmethylated cytosine (C) to uracil (U), which is eventually shown as thymine (T) in the DNA sequencing reaction. Comparing this data with untreated DNA sequencing data can identify methylated sites. However, standard bisulfite sequencing cannot distinguish 5hmC from 5mC because both of them are resistant to bisulfite treatment.
Tet-assisted bisulfite sequencing (TAB-Seq) utilizes the activity of the Ten-Eleven Translocation (TET) family protein to gradually oxidize the 5mC and eventually demethylation. Briefly, the 5mC is first oxidized to 5hmC, then to 5-formylcytosine (5fC) and finally to 5-carboxycytosine (5caC). When treated with sodium bisulfite, 5caC is converted to U like unmodified C. In TAB-Seq, the genome 5hmC is first protected by glucosylation through using β-glucosyltransferase (β-GT). The DNA is then treated with TET enzymes to convert other methylated cytosines to 5caC, while keeping the glycosylated 5hmC unaffected. Finally, the DNA is treated with bisulfite. During the depth sequencing, unmethylated cytosines and 5mC are read as T, while 5hmC is still C. Combining this data with data generated from standard bisulfite sequencing yields a genome-wide 5hmC profiling at single-base resolution.
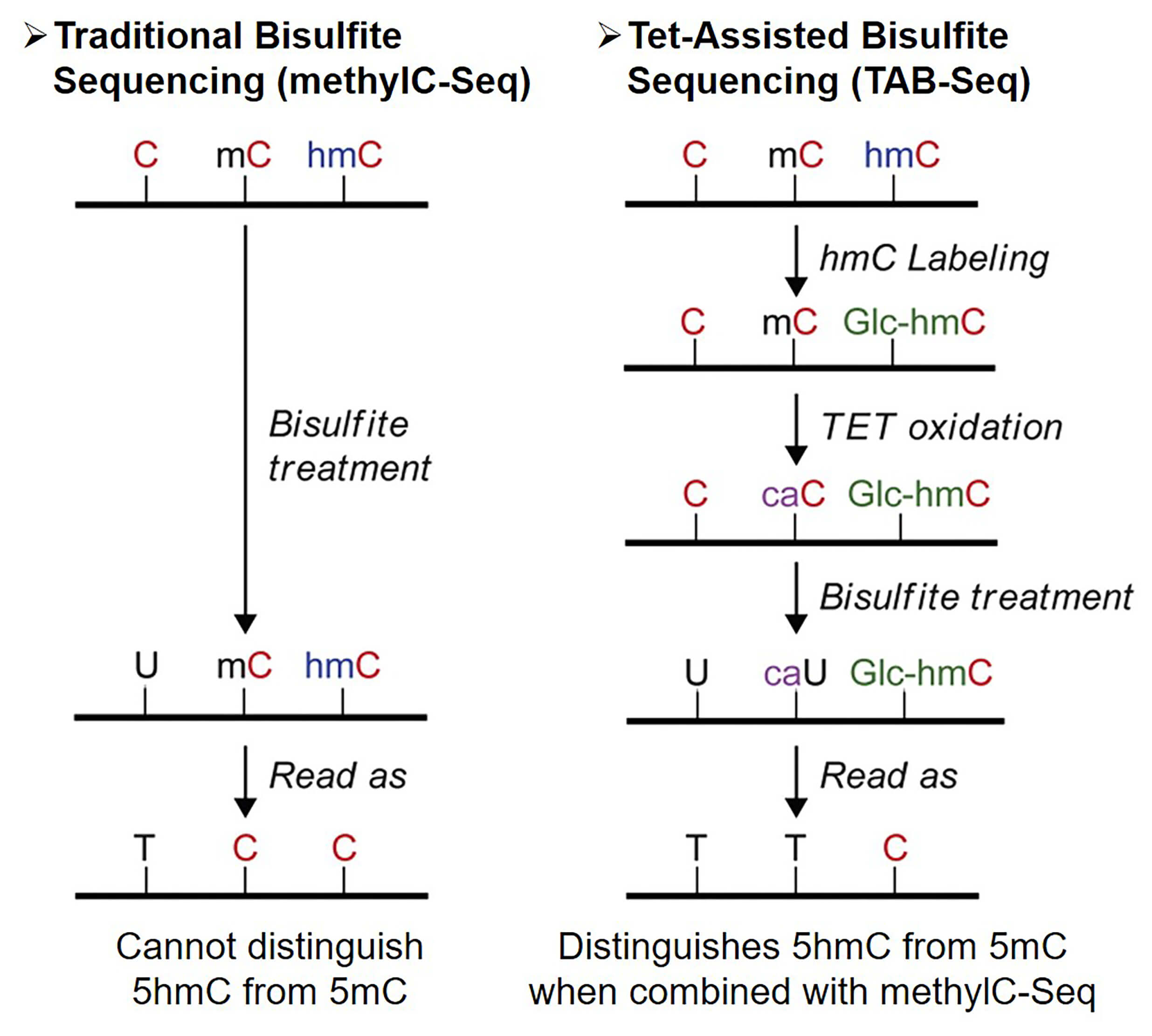
Figure 1. Overview of Tet-assisted bisulfite sequencing (TAB-Seq)
Features of Tet-Assisted Bisulfite Sequencing
Our Advantages
Workflow of Tet-Assisted Bisulfite Sequencing at Creative BioMart
Creative BioMart strictly controls each step in the TAB-Seq workflow to ensure access to reliable data quality. The inherent limitation of TAB-Seq is that it depends on the activity of the TET enzyme, and efficient conversion of 5mC to 5caC (over 96%) requires a highly active TET protein, otherwise the incomplete conversion of 5mC may lead to misidentification of the 5hmC site. As a subsidiary of Creative Biomart, a state-of-the-art protein service provider, Creative BioMart is fully capable of ensuring the activity of the TET enzyme (such as mTet1) utilized in TAB-Seq.
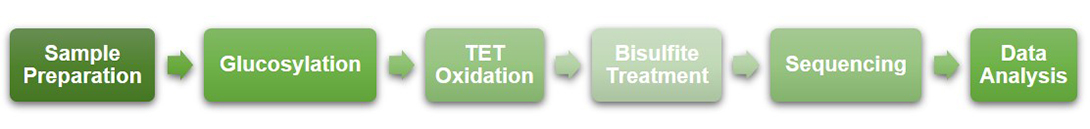
Figure 2. Workflow of TAB-Seq at Creative BioMart
Supported by our skillful experts and advanced technology platforms, Creative BioMart is dedicating to providing one-stop services for epigenetic research with high-throughput and low cost. If you have additional requirements or questions about our TAB-Seq service, please feel free to contact us.
References
1. Yu M.; et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012, 149(6): 1368.
2. Yu M.; et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nature Protocols. 2012, 7(12): 2159.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools