DNA methylation is an important epigenetic marker information, and the methylation level data of all cytosine residues in the genome-wide range is of great significance for the epigenetic space-time specificity studies. The analysis of methylation modification patterns of specific species will surely have a landmark significance in epigenomics research, and lay a foundation for the study of basic mechanisms such as cell differentiation, tissue development, animal and plant breeding, as well as human health and diseases. Based on the next-generation sequencing platform, Creative BioMart performs whole-genome bisulfite sequencing (WGBS) with low-cost, high-efficiency and high-accuracy.
What Is Whole-Genome Bisulfite Sequencing?
Whole-genome bisulfite sequencing (WGBS) is an effective and reliable strategy for the identification of methylated cytosines across the genome. This is a DNA methylation analysis method based on bisulfite treatment and sequencing. For the detection of sequences, this technique combines next-generation sequencing of high accuracy, high throughput and excellent bioinformatic analysis platform. The next-generation sequencing allows for the analysis of 5mC at single-nucleotide resolution across the genome. The principle of WGBS is to utilize bisulfite process the genome, convert the unmethylated cytosine into uracil and after PCR amplification, uracil transforms into thymine, which is distinguished from the original methylated cytosine. Combined with high-throughput sequencing technology, the methylation of CpG/CHG/CHH loci can be determined by comparing with the reference sequence. This method is capable of testing approximately 90% of the cytosines in the genomes studied so far.
Creative BioMart adopts the strategy of post-bisulfite construction to deal with the limitations of conventional WGBS. Previous methods for WGBS yielded reduced genomic representation due to required DNA shearing, ligation of methylated sequencing adapters, and bisulfite conversion of unmethylated cytosine prior to sequencing. The disadvantage of this workflow is that a large fraction of informative adaptor-tagged fragments will be degraded during bisulfite conversion and not be included in sequencing results. Our improved WGBS workflow where bisulfite conversion takes place prior to addition of sequencing adapters reduces the amount of sample, enables new types of samples to be investigated and improves CpG island coverage. At Creative BioMart, the specific library preparation method for WGBS can be chosen according to your preference, as well as our recommendation.
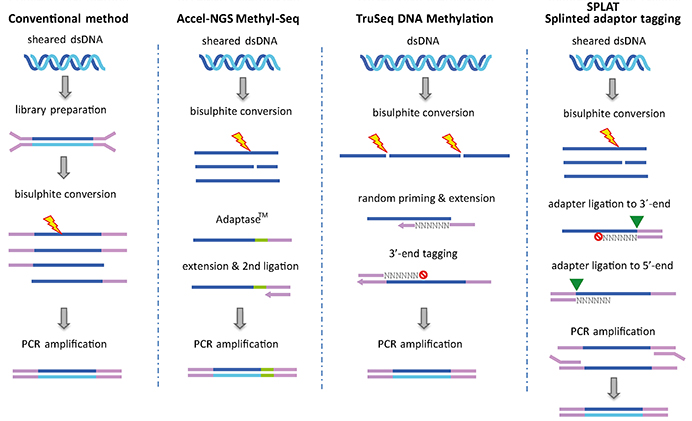
Fig 1. Principles of library preparation methods for WGBS
Features of Whole-Genome Bisulfite Sequencing
Our Advantages
Workflow of Whole-Genome Bisulfite Sequencing at Creative BioMart
Creative BioMart strictly controls each step in the WGBS workflow to ensure access to comprehensive, high-quality genome-wide methylation sequencing data. Our post-bisulfite construction strategy is that DNA samples are treated with bisulfite prior to introduction of sequencing adapters, and then sequencing.

Fig 2. Workflow of WGBS at Creative BioMart
Please contact us for more details about our whole-genome bisulfite sequencing (WGBS) service. Our experts will help design an optimal solution for your project and trouble-shoot for you throughout the whole process.
If you would like to obtain genome-wide methylation information at single-cell level, our single-cell WGBS service will meet your needs.
Reference
1. Miura, F.; et al. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Research. 2012, 40(17): e136.
2. Raine, A.; et al. SPlinted Ligation Adapter Tagging (SPLAT), a novel library preparation method for whole genome bisulphite sequencing. Nucleic Acids Research. 2016, 45(6): e36.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools