The ability to distinguish between 5mC and 5hmC is an important step toward better understanding the role of these epigenetic markers in development and disease. Many researchers believe that it is necessary to profile 5hmC at the single-nucleotide resolution, where oxidative bisulfite sequencing (oxBS-Seq) and Tet-assisted bisulfite sequencing (TAB-Seq) can achieve this goal. However, other researchers have found that region-specific or locus-specific 5hmC detection is enough to meet some research needs. And when you switch from single-nucleotide resolution to regional or locus-specific 5hmC detection, your research costs will become relatively cheap.
Creative BioMart offers hydroxymethylated DNA immunoprecipitation (hMeDIP)-based service, which is an antibody-based, large-scale technique used to enrich and capture hydroxymethylated DNA fragments for the specific region or locus in samples on a genome-wide scale. Following the hMeDIP, DNA hydroxymethylation can be analyzed using multiple downstream applications, including hMeDIP-Seq, hMeDIP-qPCR, as well as hMeDIP-chip.
What Is hMeDIP?
hMeDIP is modified from methylated DNA immunoprecipitation (MeDIP). Briefly, it utilizes 5hmC-specific antibodies to capture and enrich the hydroxymethylated DNA fragments in the genome. The resolution provided by the antibody-mediated 5hmC enrichment is determined by the resulting fragmentation profiles. Although these can vary from a few hundred to a few thousand base pairs, it is safe to say that they are best used for a more regional assessment of 5hmC abundance.
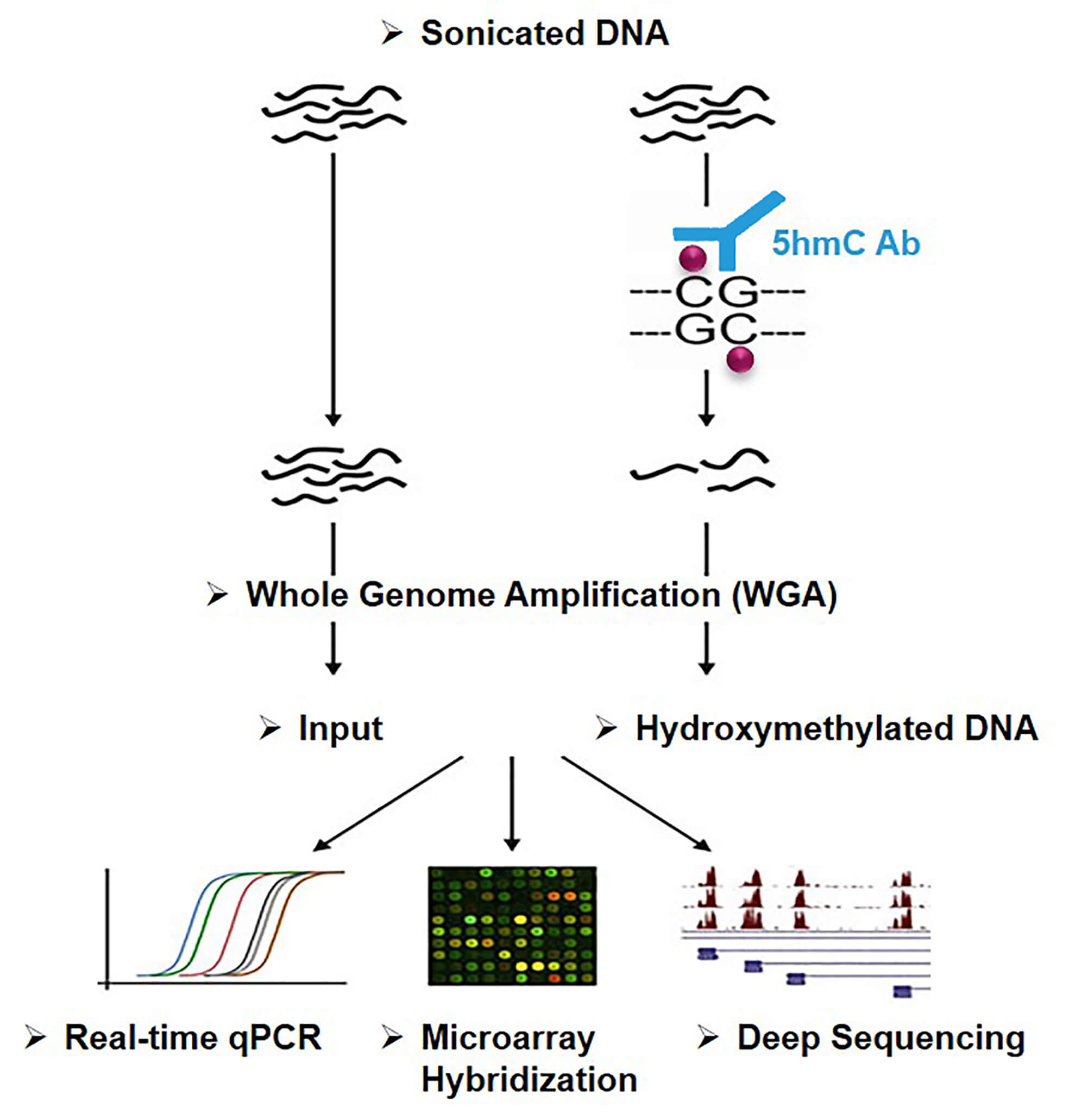
Figure 1. Principle of hMeDIP
hMeDIP-based Services at Creative BioMart

hMeDIP-Seq Service
At Creative BioMart, we combine the 5hmC monoclonal antibody capture approach with high-throughput sequencing and bioinformatics analysis to obtain a genome-wide hydroxymethylation profile. Deep sequencing provides greater genomic coverage, representing the majority of the immunoprecipitated hydroxymethylated DNA.
hMeDIP-qPCR Service
Creative BioMart perfectly combines real-time quantitative PCR technology with hMeDIP. First, the hydroxymethylated DNA fragments are enriched by hMeDIP and then detected by real-time quantitative PCR. Finally, exact values of hydroxymethylation levels of the specific region or locus in samples are obtained through data analysis.

hMeDIP-chip Service
The hMeDIP-chip is a high-throughput approach for detecting DNA hydroxymethylation across the entire genome. The hMeDIP enriched hydroxymethylated DNA fragments can be interrogated using microarrays. This combined approach allows researchers to rapidly identify hydroxymethylated regions in a genome-wide manner.
With a seasoned technical team, Creative BioMart can provide hMeDIP-based services with high-quality and efficiently, ensuring the objectives are met and accelerate your epigenetic research. Please don’t hesitate to contact us and start your project with Creative BioMart.
Reference
1. Nestor C E.; Meehan R R. Hydroxymethylated DNA immunoprecipitation (hMeDIP). Functional Analysis of DNA and Chromatin. Humana Press, 2014.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools