In mammals, 5-methylcytosine (5mC) can be converted to 5-hydroxymethylcytosine (5hmC) through the Ten-Eleven Translocation (TET) enzyme, while the traditional bisulfite sequencing approach cannot distinguish between 5mC and 5hmC. Therefore, the result of traditional bisulfite sequencing is actually a mixed signal of 5mC and 5hmC. At Creative BioMart, our oxidative bisulfite sequencing (oxBS-Seq) service can help you eliminate interference from 5hmC to achieve accurate DNA methylation detection or quantify the level of 5hmC by comparing data from bisulfite sequencing (identifies 5hmC and 5mC) to data from oxBS-Seq (identifies 5mC).
What Is Oxidative Bisulfite Sequencing?
The oxidative bisulfite sequencing (oxBS-Seq) technique is an improved bisulfite sequencing technique. OxBS-Seq differentiates 5mC and 5hmC by transforming 5hmC into 5-formylcytosine (5fC) with highly selective chemical oxidation. Unlike 5mC and 5hmC, 5fC is sensitive to the deamination of bisulfite, therefore, the cytosine (C) and 5fC of oxidized DNA are converted to uracil (U) after bisulfite treatment and read as thymine (T) at the sequencing stage. Thus, using next-generation sequencing (NGS), the cytosine (C) retained in the sequence is 5mC instead of 5hmC. Standard bisulfite conversion is performed in parallel, and the presence of 5hmC can be inferred by sequencing the oxidized and unoxidized samples. This technique is the first to conduct quantitative sequencing of 5hmC with single-base resolution and can accurately locate the location of 5hmC and 5mC. The main advantage of oxBS-Seq over enzymatic approaches such as Tet-assisted bisulfite sequencing (TAB-Seq) is that oxBS-Seq does not require highly active TET enzymes.
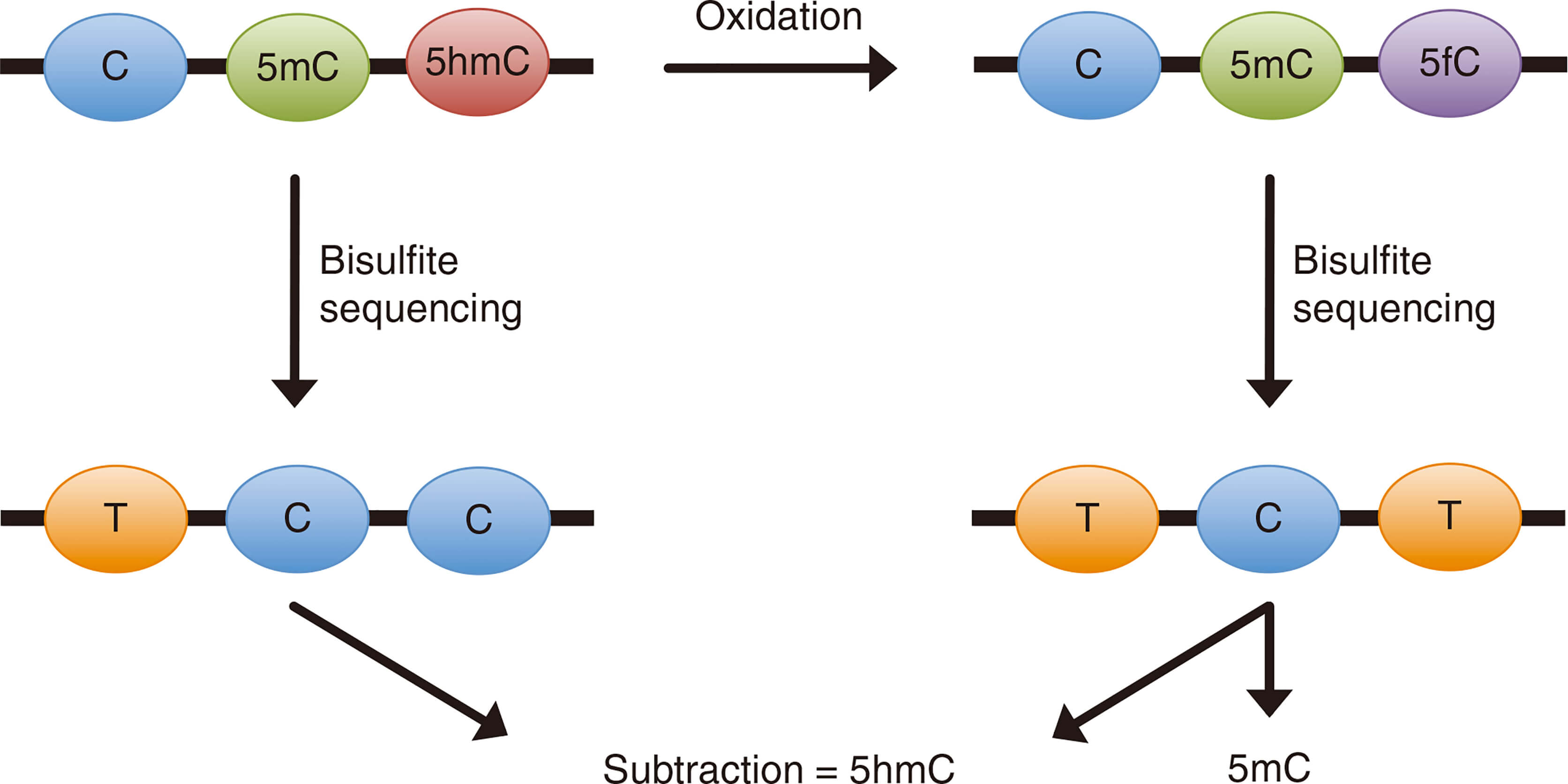
Figure 1. Schematic illustration of the principle behind oxBS-Seq (Booth M J.; et al. 2013)
Features of Oxidative Bisulfite Sequencing
Our Advantages
Workflow of Oxidative Bisulfite Sequencing at Creative BioMart
At Creative BioMart, we integrate adaptation based multiplexing and high-efficiency library preparation into the oxBS-Seq workflow to identify 5hmC levels genome-wide with the reduction of starting amount and cost per sample. Moreover, our long-term experience and advanced platforms enable us to offer a comprehensive service package from project consultation, bisulfite sequencing, oxBS-Seq and bioinformatics analysis.
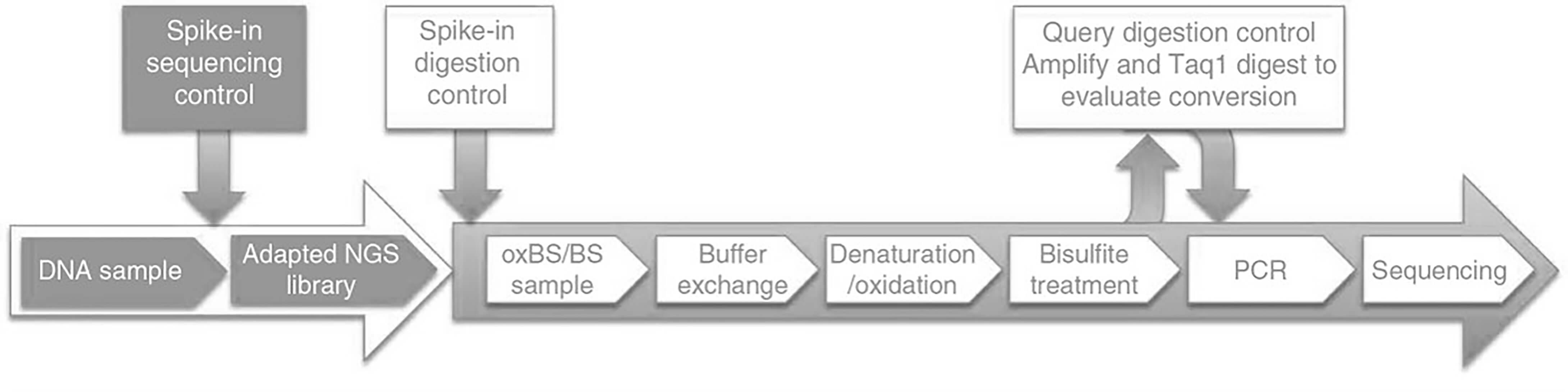
Figure 2. Workflow of oxBS-Seq at Creative BioMart
Our highly professional team in state-of-the-art labs will customize the most optimal solution to ensure your project advances quickly and promise a successful progression in epigenetic research with high quality, reliability and efficiency. Please contact us now and start your project!
References
1. Booth M J.; et al. Quantitative Sequencing of 5-Methylcytosine and 5-Hydroxymethylcytosine at Single-Base Resolution. Science. 2012, 336(6083): 934-937.
2. Booth M J.; et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nature Protocols. 2013, 8(10): 1841-1851.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools