Epigenetic status is important in transcriptional regulation, and because of the presence of different subpopulations of cells, epigenetic status in relatively homogeneous cell type may be heterogeneous, particularly in tumors, where the epigenome of individual cells is heterogeneous. Additionally, in some cases, it is difficult to obtain large numbers of cells for epigenome analysis of early-stage mammalian embryos. The single-cell reduced representation bisulfite sequencing (scRRBS) service provided by Creative BioMart can analyze the complexity of DNA methylome landscapes within a single cell.
What Is Single-cell RRBS?
The single-cell reduced representation bisulfite sequencing (scRRBS) is a methylome analysis technique that enables single-cell and single-base resolution DNA methylation analysis based on reduced representation bisulfite sequencing (RRBS). Conventional RRBS requires relatively large numbers of genomic DNA as starting material, and is not applicable to single cells. In scRRBS, all the experimental steps before PCR amplification are integrated into a single-tube reaction to minimize sample loss resulting from multiple purification steps during the workflow of RRBS. The scRRBS technique is sensitive, and capable of providing digitized methylation information on ~1 million CpG loci within an individual diploid mouse or human cell at single-nucleotide resolution. Compared with the single-cell whole-genome bisulfite sequencing (scWGBS) technique, scRRBS covers fewer CpG loci, but it provides better coverage for CpG islands, which are probably the most informative elements for DNA methylation.
Features of Single-cell RRBS
Our Advantages
Workflow of Single-cell RRBS at Creative BioMart
The basic workflow of scRRBS is as follows. Creative BioMart integrates cell lysis, methylation-insensitive restriction enzymes digestion (typically MspI), end-repair, A-tailing, adapter ligation and bisulfite conversion into a single-tube reaction to avoid unnecessary DNA loss.
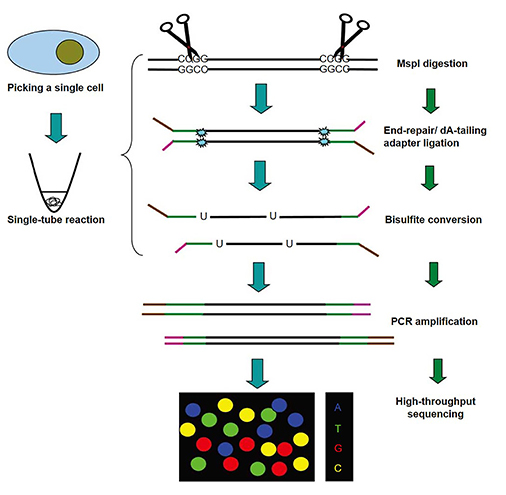
Fig 1. The schematic diagram of scRRBS
Creative BioMart strictly controls each step in the scRRBS workflow to ensure access to high quality data. Through methylation insensitive restriction enzymes combination, the enriched sequences can be more evenly distributed in regions such as promoters, CpG islands, enhancers and introns.

Fig 2. Workflow of scRRBS at Creative BioMart
Scientists at Creative BioMart are here to serve each specific need from our customers and we are more than pleased to solve every problem in the experiment to speed up your research. please don’t hesitate to contact us for more information.
Reference
1. Guo H.; et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Research. 2013, 23(12): 2126-2135.
2. Guo H.; et al. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nature Protocols. 2015, 10(5): 645-659.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools