DNA methylation plays a crucial role in epigenetic gene regulation in normal development and disease pathogenesis. Efficient and accurate quantification of DNA methylation at single-base resolution can greatly improve knowledge of disease mechanisms and be utilized to identify potential biomarkers. DNA methylation is meaningful in some genomic contexts such as CpG islands, which are found in 60% of gene promoters, and the function of DNA methylation is likely to vary depending on context. Creative BioMart provides reduced representation bisulfite sequencing (RRBS) that allows for the assessment of DNA methylation in particular genomic contexts across the genome.
What Is Reduced Representation Bisulfite Sequencing?
Reduced representation bisulfite sequencing (RRBS) is a cost-effective technique for quantification of genome-wide DNA methylation at single-base resolution. It is also a DNA methylation analysis solution based on bisulfite sequencing. In this method, CpG regions are enriched with restriction endonucleases that are insensitive to methylation, thereby reducing the amount of sequencing required while capturing most promoters and other related genome regions. RRBS allows researchers to select desired genomic contexts by choosing the density of CpG loci in the DNA regions that are examined. RRBS may be the preferred method when large numbers of samples need to be analyzed. RRBS brings down the scale and cost of whole-genome bisulfite sequencing (WGBS) by only sequencing a reduced, representative sample of the entire genome.
Features of Reduced Representation Bisulfite Sequencing
Our Advantages
Workflow of Reduced Representation Bisulfite Sequencing at Creative BioMart
The basic workflow of RRBS is as follows. Purified genomic DNA is digested by the methylation-insensitive restriction enzymes (typically MspI) to generate short fragments that contain CpG loci at the ends. After end-repair, A-tailing and ligation to methylated adapters, the CpG-rich DNA fragments are size selected, subjected to bisulfite conversion, PCR amplified and then sequenced.
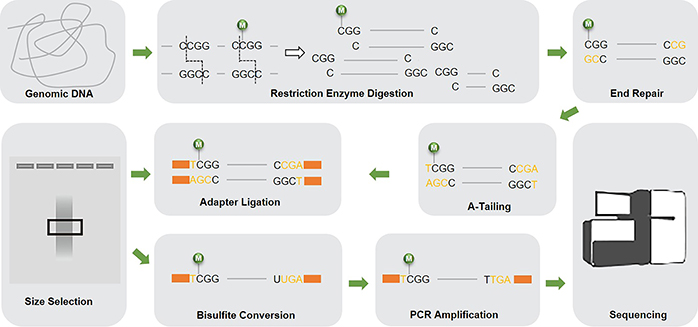
Fig 1. The schematic diagram of RRBS
Creative BioMart strictly controls each step in the RRBS workflow to ensure access to unsurpassed data quality. Through methylation insensitive restriction enzymes combination, the enriched sequences can be distributed more evenly in regions such as promoters, CpG islands, enhancers, and introns.

Fig 2. Workflow of RRBS at Creative BioMart
Creative BioMart offers reduced representation bisulfite sequencing (RRBS) service with high-throughput and low cost, if you have any questions about this service, please feel free to contact us.
If you would like to carry out reduced representation bisulfite sequencing at the single-cell level, our single-cell RRBS service will meet your needs.
Reference
1. Meissner, A.; et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Research. 2005, 33(18): 5868-5877.
2. Gu H.; et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature Protocols. 2011, 6(4): 468-481.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools