Merry Christmas and Happy New Year

Merry Christmas and Happy New Year!
A sincere thanks to you for showing full faith in us and believing in us. We value our relationship, in which we achieved so much. In the coming year, we will strive to strengthen this bond even more. From the entire team at Creative BioMart, we wish you a merry Christmas and holiday season.

What we offer?
Get 10% off for all products and services on Creative BioMart
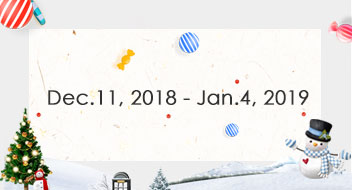
When does this discount available?
Dec.11, 2018 - Jan.4, 2019

How can I get this discount?
Please paste the code in the ‘Project Description’ area when making inquiry or purchase. Discount code: 2018Holiday10%OFF.

