Apoptosis Transcription Factors and Regulators
Related Symbol Search List
- C1D
- CITED2
- E2F1
- E2F2
- E2F4
- ETS1
- FOS
- FOSB
- FOXO3
- IRF6
- IRF8
- JUND
- MAD1L1
- MAF
- MAX
- MSX1
- MYC
- NFIL3
- PIAS3
- REL
- RELA
- RELB
- STAT1
- STAT2
- STAT3
- STAT4
- STAT6
- p53
- TRPS1
Immunology Background
Background
The execution of apoptosis involves a cascade of molecular events, primarily controlled by a group of specialized proteins known as transcription factors and regulators. These molecules orchestrate the expression of pro-apoptotic and anti-apoptotic genes, thereby determining the fate of the cell.
Transcription factors and regulators are key players in the apoptosis process, acting as molecular switches that control gene expression. They can either promote or inhibit apoptosis by binding to specific DNA sequences and regulating the transcription of target genes. The balance between pro- and anti-apoptotic signals is essential for normal cell function, and dysregulation of these factors can lead to diseases such as cancer, autoimmune disorders and neurodegenerative diseases.
Representative Apoptosis Transcription Factors and Regulators
p50
p50 is a key subunit of the NFκB complex. It is generated by proteolytic processing of the precursor protein p105. p50 lacks a transcriptional activation domain, meaning that it cannot activate gene expression on its own. Instead, p50 often forms heterodimers with other NFκB subunits, such as p65 (RelA), to regulate gene expression. The p50/RelA heterodimer is primarily involved in promoting cell survival by inducing the expression of anti-apoptotic genes such as Bcl-2 and IAPs (inhibitors of apoptosis proteins). However, p50 can also form homodimers (p50) that generally act as repressors of NFκB target genes, including those involved in cell survival. This repression can potentially contribute to apoptosis when anti-apoptotic signals are downregulated.
RELA (p65)
RelA, also known as p65, is another important subunit of the NFκB complex. It possesses a strong transcriptional activation domain that allows it to initiate target gene expression when it forms heterodimers with p50 or other NFκB subunits. RelA has a dual role in apoptosis. In most cases, RelA promotes cell survival by upregulating the expression of anti-apoptotic genes such as Bcl-2, Bcl-xL and IAPs. However, under certain conditions, RelA can also induce the expression of pro-apoptotic genes such as Fas ligand (FasL) and Bax, leading to apoptosis. The outcome often depends on the specific signaling context and the balance of other regulatory factors.
Bcl-3
Bcl-3 is an atypical member of the IκB family, which normally functions as an inhibitor of NFκB by sequestering NFκB subunits in the cytoplasm. However, Bcl-3 can act in the nucleus as a transcriptional co-activator or co-repressor, depending on its binding partners. Bcl-3 typically interacts with p50 or p52 homodimers in the nucleus to modulate transcription of NFκB target genes. Bcl-3 can promote cell survival by increasing the expression of anti-apoptotic genes. In some contexts, it can also repress pro-apoptotic genes, thereby inhibiting apoptosis. The role of Bcl-3 is often associated with tumorigenesis, where it contributes to the survival of cancer cells.
p52
p52 is another NFκB subunit generated by the processing of the precursor protein p100. Like p50, p52 lacks a transcriptional activation domain and typically forms heterodimers with other NFκB subunits such as RelB. The p52/RelB heterodimer is primarily associated with the alternative NFκB pathway, which is involved in the regulation of immune responses, lymphoid organ development, and certain aspects of cell survival. The p52/RelB complex can induce the expression of genes that promote cell survival and inhibit apoptosis, such as Bcl-2 family members. However, the role of p52 in apoptosis is context-dependent and can vary depending on the specific cellular environment and signaling inputs.
RelB
RelB is a subunit of the NFκB family that functions primarily in the alternative NFκB pathway. In contrast to RelA, RelB has a weaker transcriptional activation domain and often forms heterodimers with p52. RelB, when partnered with p52, regulates the expression of genes that can promote cell survival and inhibit apoptosis, particularly in the context of immune cells and inflammatory responses. RelB can enhance the expression of anti-apoptotic genes, thereby preventing cell death. In some cases, however, RelB has been shown to promote apoptosis, particularly in certain cancer cells, where its overexpression can lead to increased sensitivity to apoptotic signals.
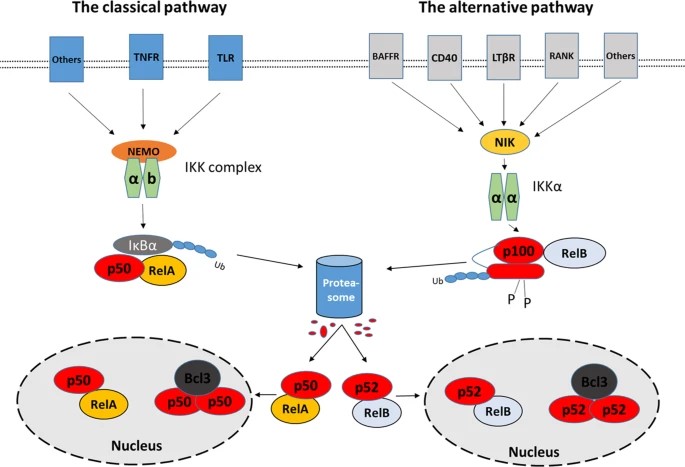 Fig. 1: Schema of the two major NFκB pathways and involved transcription factors (Dimitrakopoulos et al., 2019).
Fig. 1: Schema of the two major NFκB pathways and involved transcription factors (Dimitrakopoulos et al., 2019).STAT1 and STAT3
Signal transducer and activator of transcription (STAT) proteins are critical mediators of cytokine signaling, with STAT1 and STAT3 being prominent in the regulation of apoptosis. Ligand binding to the receptors induces receptor dimerization and activation of the receptor-associated JAK kinase, which in turn phosphorylates STAT1 and STAT3 proteins. Active STAT1/3 form a dimer and translocate to the nucleus to control gene expression. STAT1 generally promotes apoptosis, particularly in response to interferon (IFN) signaling. It induces the expression of pro-apoptotic genes such as caspases and Fas, thereby contributing to the elimination of infected or damaged cells. On the other hand, STAT3 typically acts as an anti-apoptotic factor by upregulating survival genes such as Bcl-2 and promoting survival. The balance between STAT1 and STAT3 activity can determine cell fate, with STAT1 favoring apoptosis and STAT3 promoting cell survival and proliferation.
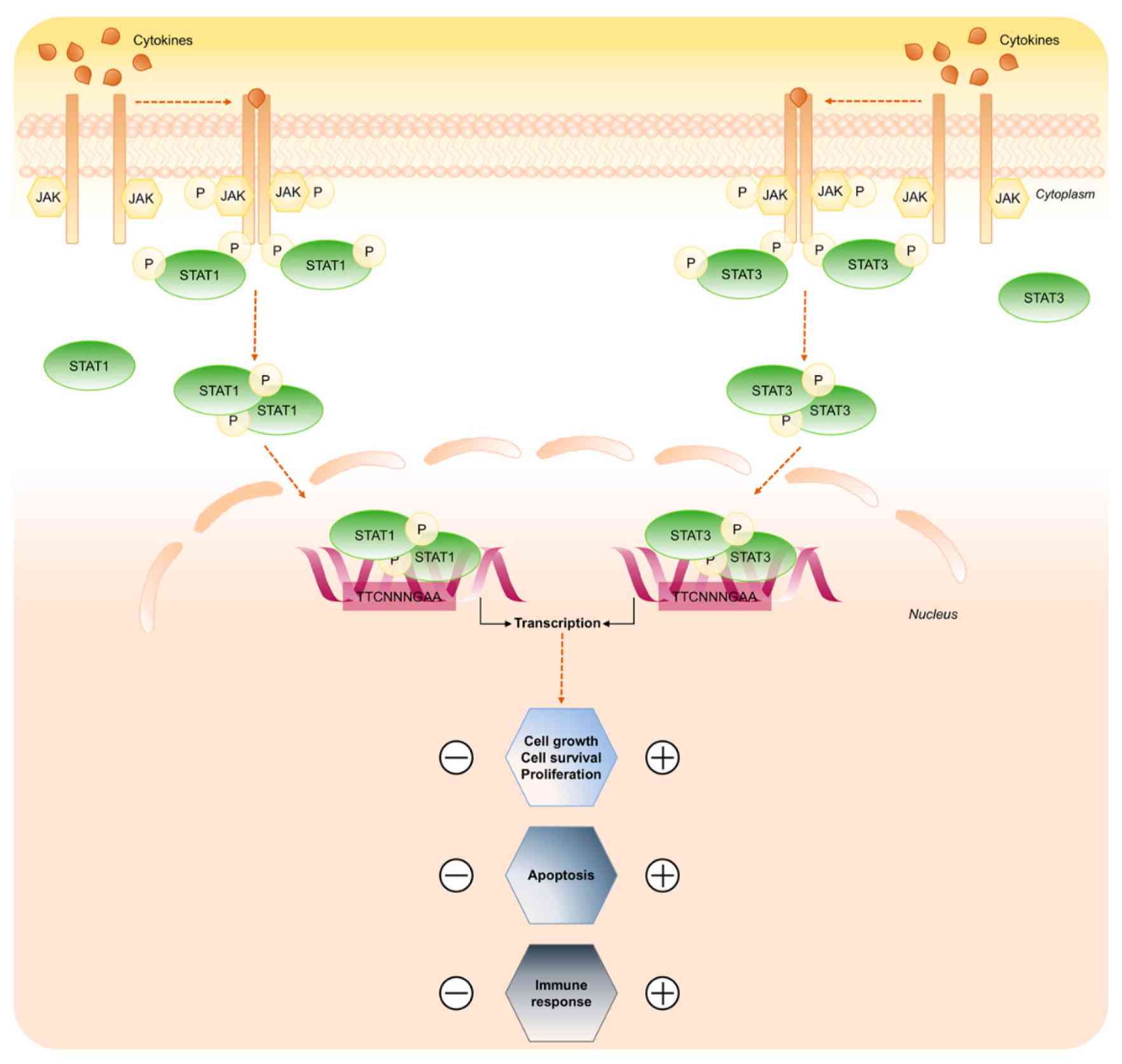 Fig. 2: STAT1 and STAT3 signaling pathway (Butturini et al., 2020).
Fig. 2: STAT1 and STAT3 signaling pathway (Butturini et al., 2020).MYC
MYC is a well-known oncogene that plays an important role in cell proliferation, growth and metabolism. Interestingly, MYC also plays a paradoxical role in apoptosis. While MYC promotes cell growth and division, it can also sensitize cells to apoptotic signals under conditions of stress or when survival factors are limited. MYC activation is a multi-step process involving multiple signaling pathways and regulatory mechanisms, including the Wnt/beta-catenin, Ras/Raf/ERK, and Ras/PI3K/AKT/GSK-3v pathways. MYC induces the expression of pro-apoptotic factors such as Bax and p53 while repressing anti-apoptotic proteins such as Bcl-2. The ability of MYC to drive both proliferation and apoptosis makes it a critical factor in cancer biology, where its overexpression can lead to uncontrolled cell growth but also to increased susceptibility to apoptotic triggers.
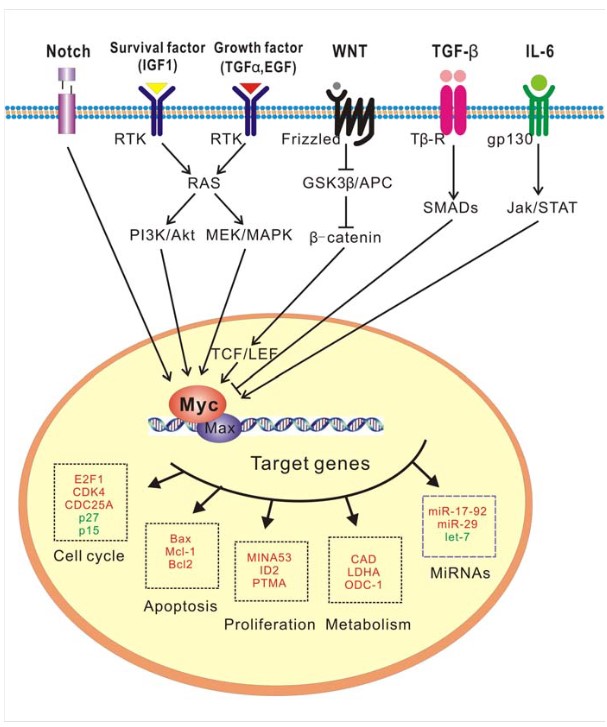 Fig. 3: A role for c-MYC as a key signaling node (Huang et al., 2014).
Fig. 3: A role for c-MYC as a key signaling node (Huang et al., 2014).FOS and JUN
FOS and JUN are components of the activator protein-1 (AP-1) transcription factor complex, which regulates a variety of cellular processes, including apoptosis. AP-1 activity is modulated by various stimuli including stress, cytokines and growth factors. Specifically, AP-1 activity is stimulated by a complex network of signaling pathways involving external signals (e.g., growth factors) and mitogen-activated protein kinases (MAPKs) of the extracellular-signal-regulated kinase (ERK), p38, and JUN-amino-terminal kinase (JNK) families. MAPKs activate various transcription factors (ternary-complex factors (TCFs), myocyte-enhancer factor 2C (MEF2C), activating transcription factor 2 (ATF2), and JUN) that induce transcription of FOS and JUN genes, thereby increasing the number of AP-1 complexes and activating AP-1 target genes. Post-translational phosphorylation by various kinases regulates AP-1 activity, including its transactivation potential, DNA binding capacity, and the stability of AP-1 components. FOS and JUN can have both pro- and anti-apoptotic roles, depending on the context. For example, AP-1 can induce apoptosis by upregulating pro-apoptotic genes such as FasL and Bim in response to cellular stress. Conversely, it can also promote cell survival by increasing the expression of anti-apoptotic genes, thereby contributing to cell proliferation and differentiation (Fig. 4).
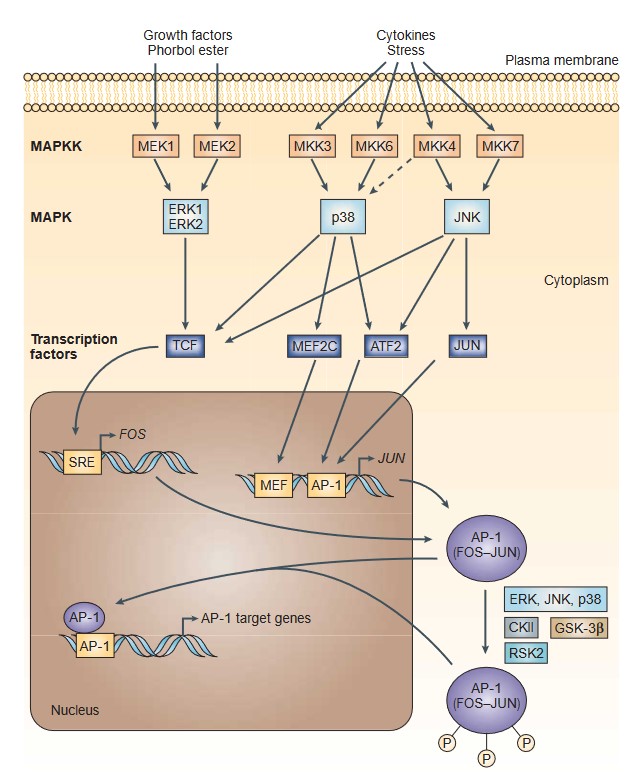 Fig. 4: A Transcriptional and post-translational activation of AP-1 (Eferl and Wagner, 2003).
Fig. 4: A Transcriptional and post-translational activation of AP-1 (Eferl and Wagner, 2003).E2F1
E2F1 is a transcription factor that plays a critical role in the regulation of the cell cycle and apoptosis. E2F1 is activated in response to DNA damage and other stress signals, leading to the transcription of genes involved in cell cycle arrest and apoptosis. E2F1 can induce apoptosis in cells with and without functional p53 protein. It transcriptionally activates genes such as ARF, ATM and Chk2, which in turn activate p53 and trigger apoptosis. However, E2F1 can also promote apoptosis independently of p53 by upregulating genes such as caspases 3, 7, 8, and 9, as well as Apaf-1, certain pro-apoptotic Bcl-2 family members, and the p53-related protein p73. Together, these activations enhance apoptotic processes. In addition, E2F1 can induce apoptosis by downregulating anti-apoptotic genes such as TRAF2 and Bcl-2.
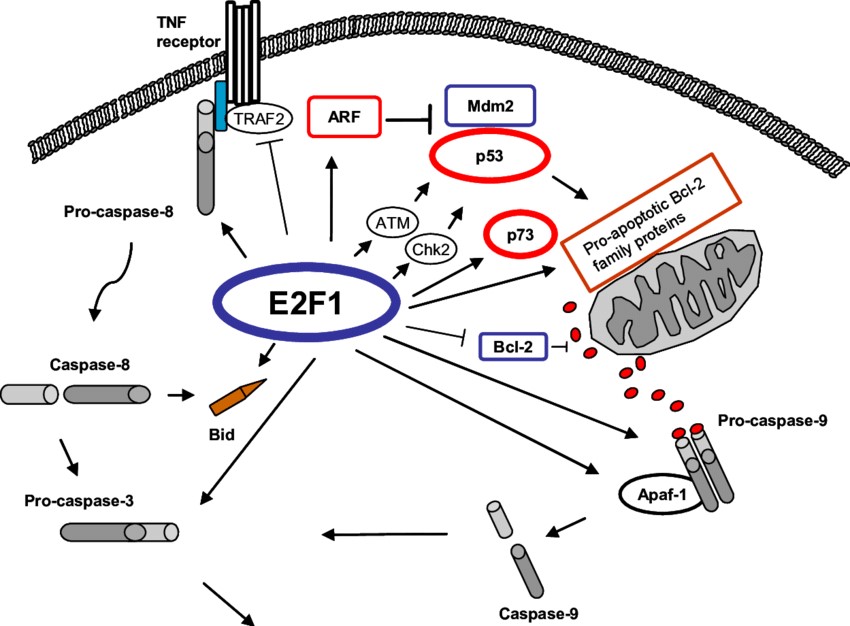 Fig. 5: E2F1 induces p53-dependent and -independent apoptosis (Helgason, 2007).
Fig. 5: E2F1 induces p53-dependent and -independent apoptosis (Helgason, 2007).Case Study
Case 1: Czuwara-Ladykowska, J.; et al. Ets1 is an effector of the transforming growth factor β (Tgf-β) signaling pathway and an antagonist of the profibrotic effects of tgf-β. Journal of Biological Chemistry, 2002; 277(23), 20399–20408.
The Ets1 transcription factor is a member of the Ets gene family and known to regulate a number of important biological processes in cells. Ets factors regulate matrix turnover by activating the transcription of several metalloproteinases (MMPs). In this study, the researchers demonstrated that Ets1 is an effector of the transforming growth factor β (TGF-β) signaling pathway and an antagonist of the profibrotic effects of TGF-β.
To determine whether Ets1 regulates ECM production in human dermal fibroblasts in the presence of TGF-β, an adenovirus expressing Ets1 was used. They evaluated the effect of Ets1 on newly synthesized collagenous proteins, including procollagens type I and III and fibronectin, and conditioned media from cells metabolically labeled with [3H]proline were analyzed by SDS-PAGE. The results showed that Ets1 expression almost completely abolished the stimulatory effect of TGF-β on collagenous protein synthesis.
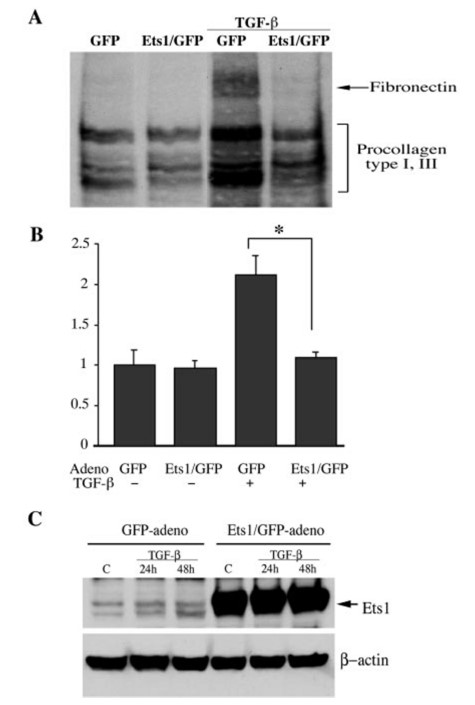 Fig. 6: Ets1 overexpression inhibits TGF-β-dependent secretion of collagenous proteins. A: representative autoradiograph. B: summary of quantitative analysis of the collagenous proteins expressed by infected fibroblasts with and without TGF-β. Bars show means ± S.E. from at least three independent experiments. The steady-state level of collagenous protein in the GFP control was arbitrarily set at 1. *, p = 0.03. C: Ets1 levels in GFP and Ets1-adenovirus-transduced fibroblasts (multiplicity of infection = 100) before and after TGF-β treatment. Blots were reprobed with anti-β-actin antibody.
Fig. 6: Ets1 overexpression inhibits TGF-β-dependent secretion of collagenous proteins. A: representative autoradiograph. B: summary of quantitative analysis of the collagenous proteins expressed by infected fibroblasts with and without TGF-β. Bars show means ± S.E. from at least three independent experiments. The steady-state level of collagenous protein in the GFP control was arbitrarily set at 1. *, p = 0.03. C: Ets1 levels in GFP and Ets1-adenovirus-transduced fibroblasts (multiplicity of infection = 100) before and after TGF-β treatment. Blots were reprobed with anti-β-actin antibody.Case 2: Erdemir, T.; et al. DNA damage-dependent interaction of the nuclear matrix protein C1D with translin-associated factor X (Trax). Journal of Cell Science, 2002; 115(1), 207–216.
C1D is a transcription factor that induces apoptosis. In addition, C1D is an activator of DNA-dependent protein kinase (DNA-PK), which is essential for DNA double-strand break (DSB) repair and V(D)J recombination. In exploring the biological function of C1D, the researchers found that C1D interacts specifically with translin-associated factor X, TRAX, using the yeast two-hybrid system. Their results suggest that one role of C1D may be to regulate TRAX/Translin complex formation.
C1D and hepatitis C virus core proteins (HCV-core/pQE) were expressed in bacteria using the plasmid pQE30 and purified by Ni-NTA column under denaturing conditions. One µg of each of the immobilized fusion proteins and pQE alone were incubated with In vitro Transcribed and Translated (IVTT) TRAX product and pull-down assay was performed. One tenth of the input TRAX was bound by C1D.
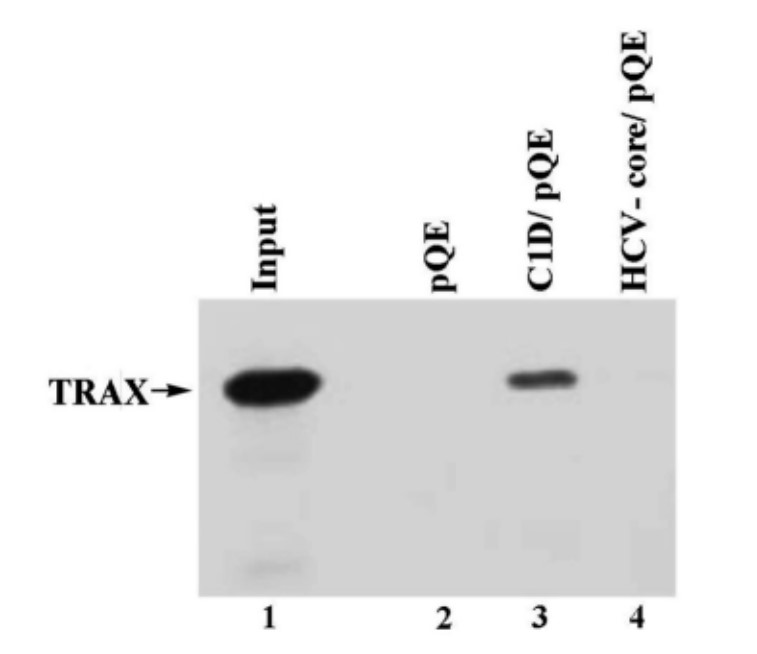 Fig. 7: C1D and TRAX interact in vitro.
Fig. 7: C1D and TRAX interact in vitro.According to the yeast two-hybrid results, TRAX binds to both C1D and Translin. Therefore, Erdemir and her colleagues wanted to determine the relative interaction between TRAX and C1D or Translin when the three molecules are found together. His-tagged C1D was immobilized on Ni-NTA beads, and in vitro transcribed and translated TRAX and Translin were incubated with the immobilized fusion protein either alone or together. The results showed that the binding of C1D and Translin to TRAX is mutually exclusive.
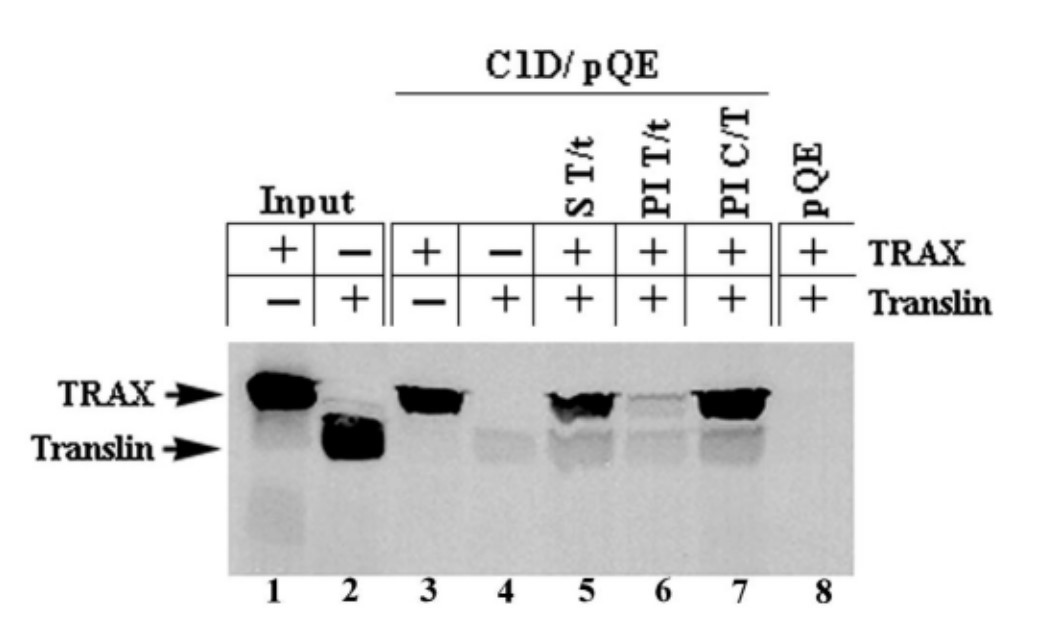 Fig. 8: Binding of C1D and Translin to TRAX is mutually exclusive. C1D was expressed in bacteria and immobilized on Ni-NTA column as explained in the legend to Fig. 7. IVTT TRAX and Translin products were incubated with the immobilized C1D either alone (lanes 3 and 4) or together. TRAX (T) and Translin (t) IVTT products were incubated simultaneously (S) with the immobilized C1D for 2 hours before washing (lane 5), TRAX and Translin IVTT products were pre-incubated (PI) for 1 hour before adding to immobilized C1D and incubated for another hour (lane 6), IVTT product of TRAX was incubated with the immobilized C1D (C) for 1 hour and then Translin was added and incubated for an additional hour (lane 7). The plasmid expressing His only (pQE) was expressed in bacteria, purified and immobilized under the same conditions as C1D and was used in lane 8 as a negative control.
Fig. 8: Binding of C1D and Translin to TRAX is mutually exclusive. C1D was expressed in bacteria and immobilized on Ni-NTA column as explained in the legend to Fig. 7. IVTT TRAX and Translin products were incubated with the immobilized C1D either alone (lanes 3 and 4) or together. TRAX (T) and Translin (t) IVTT products were incubated simultaneously (S) with the immobilized C1D for 2 hours before washing (lane 5), TRAX and Translin IVTT products were pre-incubated (PI) for 1 hour before adding to immobilized C1D and incubated for another hour (lane 6), IVTT product of TRAX was incubated with the immobilized C1D (C) for 1 hour and then Translin was added and incubated for an additional hour (lane 7). The plasmid expressing His only (pQE) was expressed in bacteria, purified and immobilized under the same conditions as C1D and was used in lane 8 as a negative control.References
- Butturini, E., Carcereri de Prati, A., & Mariotto, S. (2020). Redox regulation of stat1 and stat3 signaling. International Journal of Molecular Sciences, 21(19), 7034.
- Czuwara-Ladykowska, J., Sementchenko, V. I., Watson, D. K., & Trojanowska, M. (2002). Ets1 is an effector of the transforming growth factor β (Tgf-β) signaling pathway and an antagonist of the profibrotic effects of tgf-β. Journal of Biological Chemistry, 277(23), 20399–20408.
- Dimitrakopoulos, F.-I. D., Antonacopoulou, A. G., Kottorou, A. E., Panagopoulos, N., Kalofonou, F., Sampsonas, F., Scopa, C., Kalofonou, M., Koutras, A., Makatsoris, T., Dougenis, D., Papadaki, H., Brock, M., & Kalofonos, H. P. (2019). Expression of intracellular components of the nf-κb alternative pathway (Nf-κb2, relb, nik and bcl3) is associated with clinical outcome of nsclc patients. Scientific Reports, 9(1), 14299.
- Eferl, R., & Wagner, E. F. (2003). AP-1: A double-edged sword in tumorigenesis. Nature Reviews Cancer, 3(11), 859–868.
- Erdemir, T., Bilican, B., Oncel, D., Goding, C. R., & Yavuzer, U. (2002). DNA damage-dependent interaction of the nuclear matrix protein C1D with translin-associated factor X (Trax). Journal of Cell Science, 115(1), 207–216.
- Helgason, G. (2007). E2F1 induction following DNA damage and oncogene activation. Dissertation, University of Glasgow.
- Huang, H., Weng, H., Zhou, H., & Qu, L. (2014). Attacking c-Myc: Targeted and combined therapies for cancer. Current Pharmaceutical Design, 20(42), 6543–6554.

