Protein Deubiquitination Assay
Creative BioMart provides a reliable and efficient Protein Deubiquitination Assay designed to analyze the activity of deubiquitinating enzymes (DUBs). This assay offers a streamlined solution for assessing enzyme function in both purified recombinant proteins and cell extracts from human, mouse, and rat systems. Utilizing a fluorescent ubiquitin substrate, the assay enables sensitive detection of DUB activity with high reproducibility and flexibility, making it an ideal tool for basic research, drug discovery, and inhibitor screening.
Background: Protein Deubiquitination and Deubiquitinating Enzymes (DUBs)
Protein Deubiquitination is a critical regulatory process in eukaryotic cells that involves the removal of ubiquitin molecules from substrate proteins. This reversal mechanism is catalyzed by enzymes called deubiquitinases (DUBs), also known as deubiquitylating or ubiquitin deconjugating enzymes.
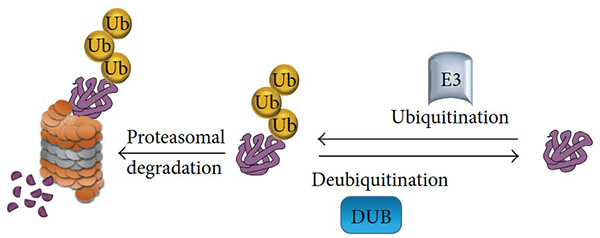
Figure 1. The ubiquitination and deubiquitination processes. A ubiquitin E3 ligase enzyme catalyzes the transfer of ubiquitin to lysine residue on the targeted protein and channels the protein to the 26S proteasome for protein degradation. Another class of enzyme, called deubiquitinating enzymes, that is able to reverse ubiquitin conjugation from protein substrates, thereby preventing proteolysis. (Suresh et al., 2016)
Major DUB Families
DUBs constitute a large family of proteases that catalyze the removal of ubiquitin from target proteins. The human genome encodes nearly 100 DUBs, making them the most extensive group of enzymes within the ubiquitin system.
| DUB Family |
Examples |
Specificity/Linked Diseases |
|---|---|---|
|
Ubiquitin-Specific Proteases (USPs) |
Broad specificity; cancer, neurodegeneration |
|
|
Ovarian Tumor Proteases (OTUs) |
Immune regulation, inflammation |
|
|
JAMM/MPN+ Metalloproteases |
PSMD14, AMSH |
Cleaves K63 chains; DNA repair, endocytosis |
|
Josephin Domain DUBs |
Polyglutamine diseases (e.g., spinocerebellar ataxia) |
Key Functions
These enzymes play pivotal roles in:
- Ubiquitin precursor processing
- Ubiquitin recycling
- Trimming ubiquitin chains
- Regulation of cell growth and differentiation
- DNA damage repair and disease pathways
- Transcriptional regulation and chromatin remodeling
Because of their central role in cellular homeostasis and disease mechanisms, DUBs have become highly attractive targets for biomedical research and therapeutic development.
What We Offer
Creative BioMart’s Protein Deubiquitination (DUB) Assay offers a reliable and efficient platform for measuring deubiquitination enzyme activity in human, mouse, and rat systems. This assay is designed for use with both cell extracts and purified recombinant proteins, making it suitable for a wide range of research applications. The system employs a fluorescent ubiquitin-based substrate, which generates a measurable fluorescent signal upon cleavage by cysteine protease DUBs. The resulting fluorescence is directly proportional to enzymatic activity, providing a sensitive, quantitative, and high-throughput-ready solution for DUB screening and functional studies.
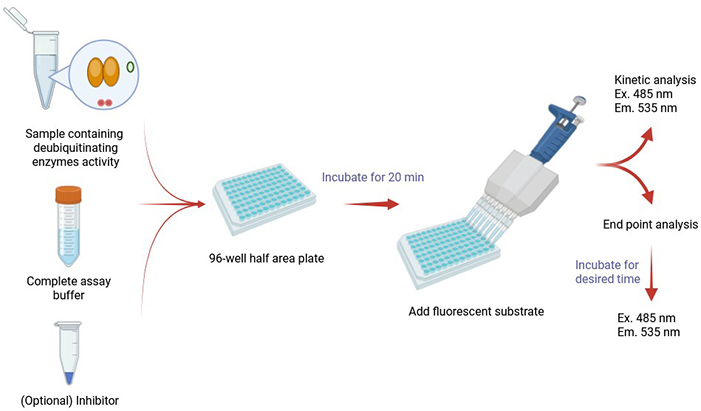
This service supports researchers in exploring the fundamental roles of DUBs while offering practical applications for drug discovery pipelines.
Service Details
|
Item |
Details |
|---|---|
|
Substrate |
Fluorescently labeled ubiquitin for sensitive activity detection |
|
Controls |
Positive control extracts and universal DUB inhibitor provided |
|
Detection Method |
Fluorescent readout at Ex/Em = 485/535 nm |
|
Applications |
|
Service Workflow

Why Partner with Us
- Comprehensive Assay Kit: Includes optimized buffers, controls, and inhibitor for reliable results.
- High Sensitivity & Specificity: Detects activity with a strong fluorescent signal and minimal background.
- Flexible Analysis: Suitable for both kinetic monitoring and endpoint measurements.
- Cross-Species Compatibility: Supports human, mouse, and rat protein systems.
- Drug Discovery Utility: Facilitates rapid and efficient screening of DUB inhibitors.
- Expert Support: Our experienced team provides consultation and customized solutions for unique research needs.
Case Studies and Applications
Case 1: Fluorescent assay for monitoring deubiquitinating enzymes
Hassiepen et al., 2007. doi:10.1016/j.ab.2007.07.034
Ubiquitin modification is a critical regulatory mechanism in eukaryotic cells, governing protein stability, localization, and function. To study its reversal, a robust fluorescence-based assay has been developed for monitoring the activity of human deubiquitinating proteases, including USP2 and UCH-L3. Using the novel substrate ubiquitin-rhodamine110-glycine, the assay offers a high dynamic range and excellent optical properties, enabling precise kinetic characterization. Optimized for 384-well format, it achieves remarkable sensitivity with protease concentrations as low as 120 pM (USP2) and 1 pM (UCH-L3). This system is ideally suited for inhibitor screening and allows determination of inhibitory constants in the subnanomolar range.
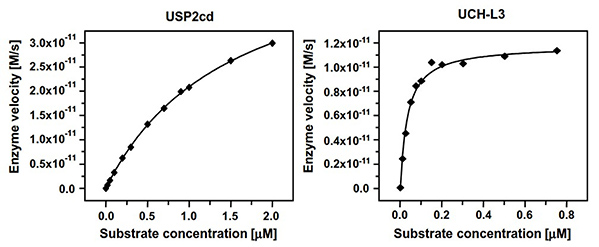
Figure 2. Determination of Km and kcat of Ub-Rho110-G for USP2cd and UCH-L3. Assays were conducted for USP2cd and UCH-L3 in 384-well plates. (Hassiepen et al., 2007)
Case 2: Multiplex assays for deubiquitinating enzyme inhibitor discovery
Tian et al., 2011. doi:10.1089/adt.2010.0317
The reversible conjugation of ubiquitin and ubiquitin-like proteins is vital for regulating diverse cellular pathways, with deubiquitylases (DUBs) playing a central role by removing these modifications. Given their substrate specificity and involvement in signaling, DUBs are promising therapeutic targets. To accelerate drug discovery, the researchers developed two novel assays—UbL-Enterokinase light chain and UbL-Granzyme B—complementing an established assay in a multiplex format. This system enables simultaneous measurement of three protease activities in a single well, improving efficiency and selectivity assessment. The multiplex assay successfully distinguished selective inhibitors, including characterization of P022077, a USP7-specific inhibitor.
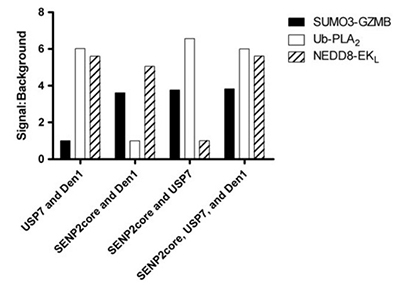
Figure 3. SUMO3-GZMB, ubiquitin-PLA2 , and NEDD8-EKL were incubated with either the complete mixture containing SENP2 core, USP7, and DEN1, or a mixture lacking one of the three isopeptidases. Fluorescence intensity was measured following a 60-minute incubation and signal:background was calculated. (Tian et al., 2011)
Client Success Stories
"We engaged Creative BioMart’s Protein Deubiquitination Assay service to evaluate DUB activity in cancer cell extracts as part of our oncology drug discovery program. Their assay identified elevated DUB activity in tumor-derived samples compared to healthy tissue, helping us validate our therapeutic target. The inclusion of a universal DUB inhibitor provided a reliable benchmark, and the detailed kinetic data gave our team the confidence to prioritize lead compounds for further development."
— Senior Scientist, Oncology Research | Mid-Sized Biotech Company
"Our pharmaceutical team needed a robust platform to screen a library of small molecules for potential DUB inhibitors. Creative BioMart’s fluorescent-based assay delivered exactly what we needed: rapid, reproducible, and high-sensitivity results. The optimized buffers ensured consistent enzyme activity, and the data reports were thorough enough to move several hits into our preclinical testing pipeline. This collaboration significantly shortened our early-stage discovery timeline."
— Director of Drug Discovery | Global Pharmaceutical Company
"Our academic lab studies ubiquitin biology in neurodegeneration, and we required an assay that could compare DUB activity across human, mouse, and rat models. Creative BioMart’s service provided a clear readout of enzymatic activity across all three systems, confirming the conservation of DUB regulation in our disease model. The assay’s flexibility for both kinetic and endpoint analyses gave us valuable mechanistic insights."
— Principal Investigator, Molecular Biology | Leading Research University
"In our epigenetics research program, we were investigating chromatin remodeling pathways influenced by DUB activity. Creative BioMart’s Protein Deubiquitination Assay allowed us to test the effects of candidate compounds on specific histone-modifying DUBs. Their expert team even provided guidance on sample preparation to maximize signal sensitivity."
— Head of Translational Research | Independent Research Institute
FAQs About Protein Deubiquitination Assay
-
Q: What types of samples can be used with the protein deubiquitination assay?
A: Our assay is compatible with cell extracts and purified recombinant proteins from human, mouse, and rat systems. This flexibility allows you to explore DUB activity across different biological models, from basic research to preclinical drug development. -
Q: How sensitive is the assay, and what detection method is used?
A: The assay uses a fluorescent ubiquitin substrate that releases a strong signal upon cleavage by DUBs. Detection is performed at an excitation wavelength of 485 nm and emission wavelength of 535 nm, ensuring high sensitivity and reproducibility with minimal background noise. -
Q: Can the assay be used for inhibitor screening?
A: Yes. The Protein Deubiquitination Assay is ideal for rapid screening of DUB inhibitors. It includes both a positive control extract and a universal DUB inhibitor, making it straightforward to compare compound effects against validated controls. -
Q: Does the service support kinetic as well as endpoint analysis?
A: Absolutely. The assay is optimized for both kinetic monitoring of enzymatic activity over time and endpoint measurements, giving you flexibility depending on your experimental design and research goals. -
Q: Can you provide customized assay solutions?
A: Yes. Our scientific team works closely with clients to tailor assay conditions, sample preparation, and analysis workflows to meet specific research needs—whether you’re studying fundamental ubiquitin biology or testing compounds for therapeutic potential.
Resources
Related Services
Related Products
References:
- Hassiepen U, Eidhoff U, Meder G, et al. A sensitive fluorescence intensity assay for deubiquitinating proteases using ubiquitin-rhodamine110-glycine as substrat e. Analytical Biochemistry. 2007;371(2):201-207. doi:10.1016/j.ab.2007.07.034
- Suresh B, Lee J, Kim KS, Ramakrishna S. The importance of ubiquitination and deubiquitination in cellular reprogramming. Ruohola-Baker HT, ed. Stem Cells International. 2016;2016(1):6705927. doi:10.1155/2016/6705927
- Tian X, Isamiddinova NS, Peroutka RJ, et al. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. ASSAY and Drug Development Technologies. 2011;9(2):165-173. doi:10.1089/adt.2010.0317
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

