Protein Network Construction and Topological Structure Analysis
Creative BioMart provides comprehensive Protein Network Construction and Topological Structure Analysis Services designed to support researchers in deciphering complex protein interactions and structural properties. With over ten thousand successful bioinformatics consultations delivered worldwide, we combine cutting-edge computational technologies with expert statistical and informatics support. Our services enable accurate prediction, visualization, and analysis of protein networks, crosslinks, and topologies across multiple species. By integrating curated databases and customized analytical strategies, Creative BioMart empowers clients to accelerate discoveries in molecular biology, drug development, and systems-level studies.
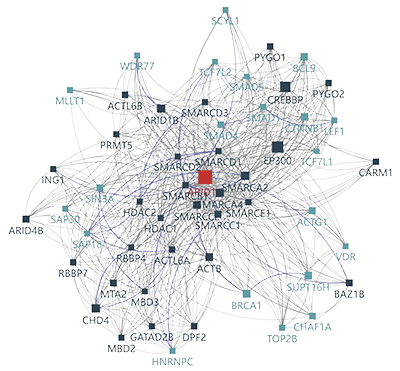
Understanding Protein Network Construction and Topological Analysis
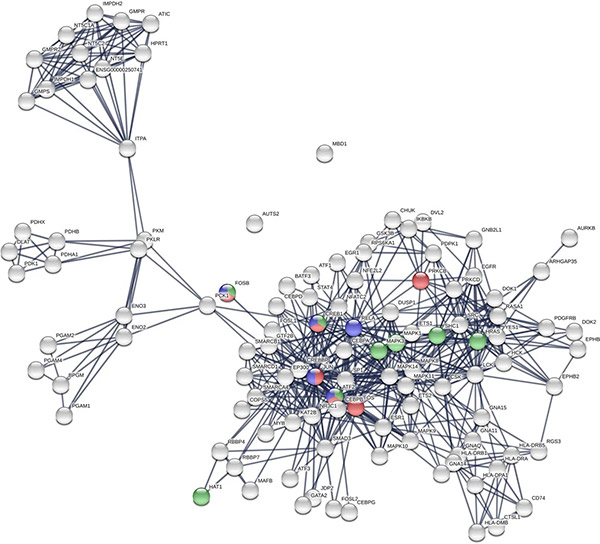
Understanding protein interactions and topological structures is central to unraveling the molecular mechanisms of life. Proteins rarely act in isolation; instead, they form dynamic interaction networks that influence biological processes, disease progression, and therapeutic outcomes. The topological properties of these networks—such as connectivity, clustering, and modular organization—provide critical insights into protein function and pathway regulation.
However, analyzing protein networks requires advanced computational methods, domain expertise, and curated datasets. Creative BioMart addresses these challenges by offering professional services in protein network construction, topology prediction, and statistical analysis, enabling researchers to focus on biological interpretation while relying on accurate, high-quality computational outputs.
Protein Network and Topological Analysis: Our Advanced Services
Service Workflow
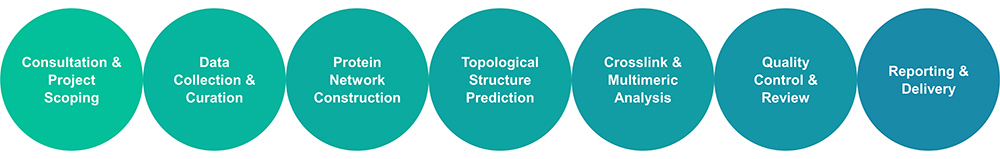
Our Services
Creative BioMart delivers a full spectrum of services for protein network analysis and topological structure prediction. Our team provides customized assistance in:
-
De Novo Tandem Repeat Detection
Detect tandem repeats in protein sequences to aid studies of folding, stability, and molecular interactions.
-
Protein Crosslink Computation
Compute intra- and inter-protein crosslinks to support structural analysis and characterization of protein assemblies.
-
Sequence Similarity Search
Perform sensitive searches in curated databases to identify homologs, annotate functions, and explore phylogenetics.
-
Membrane Protein Topology Prediction
Predict transmembrane helices, signal peptides, and orientation to guide structural and functional studies of membrane proteins.
-
Molecular Topology Consultation
Provide guidance on connectivity, stereochemistry, and physicochemical properties for ligand design and docking studies.
-
Asymmetric vs. Biological Unit Comparison
Compare asymmetric and biological units to clarify quaternary structures and functional states in crystals.
-
Multimeric Molecule Reconstruction
Reconstruct multimeric assemblies from crystallographic data to study protein-protein interactions and complex mechanisms.
Supported Species
Additionally, we support manual review and annotation for species including mouse, rat, Xenopus laevis, Xenopus tropicalis, and zebrafish, alongside the human proteome and other mammalian chordates. This makes our services especially valuable for embryonic development and cell biology studies using key model organisms.




Service Details
- Core Features: De novo repeat detection, topology prediction, signal peptide annotation, crosslink mapping, asymmetric unit comparison, and multimeric molecule reconstruction.
- Databases & Tools: Integration with curated model organism databases and species-specific resources.
- Customization: Tailored workflows and datasets to meet client-specific needs in developmental biology, structural biology, and drug discovery.
- Deliverables: Comprehensive analytical reports with visualizations, annotations, and recommendations for experimental validation.
Why Partner with Creative BioMart
- Proven Expertise: Over 10,000 successful bioinformatics consultations, serving researchers worldwide.
- Comprehensive Services: From protein crosslink mapping to membrane protein topology prediction, all in one place.
- Species-Specific Insights: Specialized support for model organisms ( Xenopus , zebrafish, rodents) alongside human proteome analysis.
- Advanced Infrastructure: Bioinformatics core facilities equipped with cutting-edge computational tools and curated resources.
- Quality Assurance: Rigorous statistical support, manual curation, and collaboration with trusted model organism databases.
- Customizable Solutions: Flexible, project-specific workflows tailored to both academic and industrial research needs.
Representative Case Studies
Case 1: Identification of COPD blood-based biomarkers through network analysis
Banaganapalli et al., 2022. doi:10.1371/journal.pone.0274629
Chronic obstructive pulmonary disease (COPD) is a progressive respiratory disorder with significant global morbidity and mortality. This study used computational analysis of microarray gene expression datasets from blood and lung tissues to identify shared differentially expressed genes (DEGs). A total of 63 DEGs were found, from which 12 hub gene-network clusters (including SREK1, TMEM67, IRAK2, MECOM, and ASB4 ) were prioritized through protein network construction and functional enrichment analysis. These genes were linked to inflammation, immune activity, and airway remodeling. Several, such as IRAK2 and MECOM, were also implicated in pulmonary diseases and lung cancer, highlighting their biomarker potential.
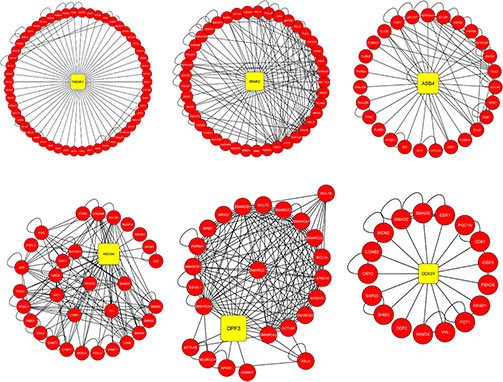
Figure 1. Hub genes TMEM67, IRAK2, ASB4, MECOM, DPF3 and DDX3Y with their clusters identified from the common DEGs between blood and lung tissue datasets. (Banaganapalli et al., 2022)
Case 2: Improved detection of protein complexes using link-based clustering
Hu et al., 2021. doi:10.1109/TNSE.2021.3109880
Protein complexes are essential functional modules that drive cellular organization and processes, making their accurate detection critical for biological research. Traditional clustering algorithms often rely on protein-level criteria, overlooking the fact that proteins operate through interactions. To address this, a novel link-based clustering algorithm was developed, focusing on pairwise interaction similarities derived from network topology and Gene Ontology. By formulating the problem as an optimization task, this method effectively identifies overlapping protein complexes. Tests on five yeast and human PPI datasets demonstrated significantly higher detection accuracy compared to state-of-the-art approaches, revealing functionally meaningful complexes with improved precision.
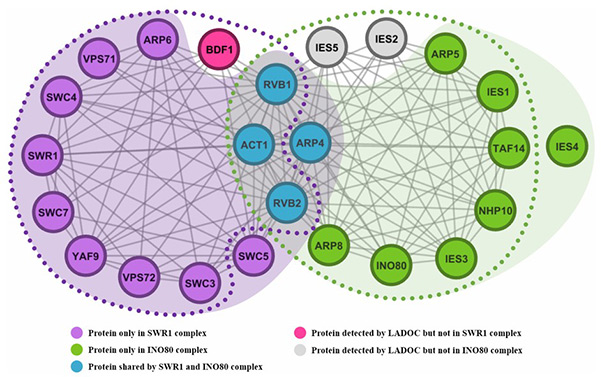
Figure 2. An example of overlapping protein complexes detected by LADOC from the PPI network of Collins. (Hu et al., 2021)
Customer Testimonials: What Our Clients Say
"We engaged Creative BioMart to construct protein interaction networks for our oncology biomarker program. Their team built a detailed map of crosslinks and pathway associations, integrating both public databases and our in-house data. The insights helped us prioritize key signaling nodes and refine our therapeutic strategy. The accuracy and depth of their network analysis exceeded expectations and gave us a solid foundation for biomarker validation."
— Director of Translational Oncology | Global Pharmaceutical Company
"Our lab struggled with predicting the topology of several novel membrane proteins implicated in ion transport. Creative BioMart delivered precise predictions of transmembrane domains and signal peptides, which guided our mutagenesis experiments. Their predictions aligned perfectly with our cryo-EM data, saving us months of work. Their ability to integrate structural prediction with biological interpretation made them a trusted collaborator on this project."
— Principal Investigator, Membrane Biology | Academic Research Institute
"In one of our crystallography projects, reconstructing the biological assembly of a protein complex proved challenging. Creative BioMart provided reconstruction of multimeric molecules from crystal data and compared asymmetric units to biological units. Their analysis clarified the quaternary structure, which directly supported our drug design pipeline. Their timely delivery and expert consultation allowed us to meet critical project deadlines with confidence."
— Head of Structural Biology | Biotechnology Company
"Our zebrafish project required reliable annotation and network construction for proteins involved in embryonic development. Creative BioMart combined de novo repeat detection, topology prediction, and manual database curation to produce a robust dataset. Their close collaboration with model organism databases ensured the accuracy of our annotations."
— Lead Scientist, Developmental Biology | Research Consortium
FAQs About Protein Network Construction and Topological Analysis
-
Q: What is protein network construction and why is it important for my research?
A: Protein network construction helps map interactions between proteins, revealing functional modules and pathways that drive cellular processes. This provides critical insights into disease mechanisms, biomarker discovery, and drug target identification. -
Q: What types of services do you offer in network construction and topology analysis?
A: We offer de novo tandem repeat detection, prediction of membrane protein topology, sequence similarity searches, crosslink computation, multimeric molecule reconstruction, and consultation on small molecule topology. Our services cover both human proteins and key model organisms such as mouse, rat, zebrafish, and Xenopus . -
Q: How accurate are your predictions and analyses?
A: Our analyses leverage advanced computational algorithms and manually curated databases, ensuring high accuracy and biological relevance. We also integrate Gene Ontology annotations and model organism resources to maximize reliability. -
Q: Can you help with both experimental data and database-driven projects?
A: Yes. Whether you provide experimental data that requires analysis or seek comprehensive database-driven predictions, we tailor our workflow to support both approaches. -
Q: What species do you support in your analysis?
A: In addition to human, we provide detailed analysis for model organisms including mouse, rat, Xenopus laevis , Xenopus tropicalis , and zebrafish. These models are essential for studies in embryonic development, cell biology, and translational research. -
Q: How does your workflow ensure reproducibility and reliability?
A: We apply standardized bioinformatics pipelines, integrate QC measures at multiple stages, and manually validate critical results against curated resources. This guarantees reproducibility across projects and platforms. -
Q: Why should I choose Creative BioMart for protein network analysis?
A: With a proven record of more than ten thousand bioinformatics consultations, Creative BioMart combines deep expertise, advanced computational platforms, and close collaborations with model organism databases. Our customized solutions are designed to accelerate research while ensuring cost-effectiveness and scientific rigor.
Resources
Related Services
References:
- Banaganapalli B, Mallah B, Alghamdi KS, et al. Integrative weighted molecular network construction from transcriptomics and genome wide association data to identify shared genetic biomarkers for COPD and lung cancer. Parine NR, ed. PLoS ONE. 2022;17(10):e0274629. doi:10.1371/journal.pone.0274629
- Chen SJ, Liao DL, Chen CH, Wang TY, Chen KC. Construction and analysis of protein-protein interaction network of heroin use disorder. Sci Rep. 2019;9(1):4980. doi:10.1038/s41598-019-41552-z
- Hu L, Zhang J, Pan X, Luo X, Yuan H. An effective link-based clustering algorithm for detecting overlapping protein complexes in protein-protein interaction networks. IEEE Trans Netw Sci Eng. 2021;8(4):3275-3289. doi:10.1109/TNSE.2021.3109880
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

