Histone Modifying Enzymes
Creative BioMart Histone Modifying Enzymes Product List
Immunology Background
Background
Histone-modifying enzymes play a crucial role in regulating gene expression and chromatin structure, thereby influencing various cellular processes such as development, differentiation, and disease. These enzymes catalyze post-translational modifications (PTMs) on histone proteins, which are the building blocks of chromatin.
Histones are proteins around which DNA is wrapped to form a compact structure called chromatin. The N-terminal tails of histones protrude from the nucleosome and can undergo various modifications, including acetylation, methylation, phosphorylation, ubiquitination, and more. Each of these modifications can have distinct effects on gene expression and chromatin accessibility.
Histone-modifying enzymes are responsible for adding or removing these PTMs. The addition or removal of PTMs by these enzymes can alter the structure and function of chromatin, leading to changes in gene expression. For example, acetylation of histones by HATs generally promotes transcription by loosening chromatin structure, while deacetylation by HDACs can have a repressive effect. Methylation of histones by HMTs can either activate or repress gene expression, depending on the specific site and context.
In summary, histone-modifying enzymes are key regulators of gene expression and chromatin structure through the addition or removal of PTMs on histone proteins. They play a fundamental role in cellular processes and have implications in various diseases. Investigating these enzymes provides insights into the mechanisms of gene regulation and opens avenues for targeted therapies.
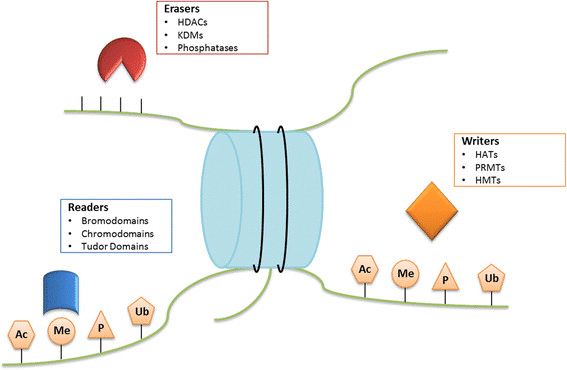 Fig.1 Writers, erasers, and readers. (Graça, I., et al., 2016)
Fig.1 Writers, erasers, and readers. (Graça, I., et al., 2016)Epigenetic Writers (HATs, HDMs, and PRMTs) are responsible to establish epigenetic marks on amino acid residues of histone tails. Epigenetic Erasers (HDACs, KDMs, and phosphatases) participate on the removal of the epigenetic marks. Epigenetic Readers (bromodomain, chromodomain, and Tudor domain proteins) recognize and bind to a specific epigenetically modified mark.
The Major Classes of Histone Modifying Enzymes
Histone-modifying enzymes are responsible for adding or removing these PTMs. They can be broadly categorized into two major groups: writers and erasers. Writers include enzymes that add PTMs, such as histone acetyltransferases (HATs), histone methyltransferases (HMTs), and kinases, while erasers encompass enzymes that remove PTMs, such as histone deacetylases (HDACs), histone demethylases (HDMs), and phosphatases. Here are some of the major classes of histone-modifying enzymes and their functions:
| Different Categories | Functions | |
|---|---|---|
| Writers | Histone Acetyltransferases (HATs) | HATs add acetyl groups to specific lysine residues on histone tails, leading to a relaxed chromatin structure and transcriptional activation. They are involved in gene expression regulation, DNA repair, and other cellular processes. |
| Histone Methyltransferases (HMTs) | HMTs add methyl groups to specific lysine or arginine residues on histones. Methylation can have diverse effects depending on the specific residue and degree of methylation. It can be associated with both gene activation and repression, and plays a critical role in chromatin organization and gene regulation. | |
| Histone Kinase | These enzymes phosphorylate specific serine or threonine residues on histones, influencing chromatin structure and dynamics. Phosphorylation of histones is often associated with transcriptional activation and other cellular processes. | |
| Erasers | Histone Deacetylases (HDACs) | HDACs remove acetyl groups from lysine residues on histones, leading to a more compact chromatin structure and transcriptional repression. They are involved in gene silencing, cell cycle regulation, and other cellular processes. |
| Histone Demethylases (HDMs) | HDMs remove methyl groups from specific lysine or arginine residues on histones, reversing the effects of methylation. They play a crucial role in maintaining proper gene expression patterns and chromatin dynamics. | |
| Histone Phosphatases | These enzymes remove phosphate groups from phosphorylated histones, regulating chromatin structure and transcriptional activity. They are involved in diverse cellular processes, including DNA repair and cell cycle control. | |
It's important to note that histone modifying enzymes often work in complex regulatory networks and can have overlapping or context-dependent functions. They play critical roles in gene regulation, chromatin remodeling, and the maintenance of cellular homeostasis. Dysregulation of these enzymes can contribute to various diseases, highlighting their significance as potential therapeutic targets.
Histone Modifying Enzymes Involved in Biological Processes
Histone modifying enzymes are involved in a wide range of biological processes due to their role in regulating gene expression and chromatin structure. Here are some of the key biological processes in which histone modifying enzymes participate:
| Biological Processes | Details |
|---|---|
| 1. Gene Expression Regulation |
Histone modifications play a fundamental role in the regulation of gene expression. By adding or removing specific modifications, histone modifying enzymes can influence the accessibility of DNA to transcription factors and the transcriptional machinery, thereby controlling the activation or repression of genes.
|
| 2. Development and Differentiation | Histone modifications are essential for embryonic development, cell lineage commitment, and cellular differentiation. They contribute to the establishment of specific gene expression programs that guide the formation and function of different tissues and cell types during development. |
| 3. Epigenetic Inheritance | Histone modifications can be stably transmitted through cell divisions, leading to the inheritance of gene expression patterns. This process, known as epigenetic inheritance, allows cells to maintain their differentiated states and ensures the stability of cellular identity across generations. |
| 4. DNA Repair and Genome Stability | Histone modifying enzymes participate in DNA repair processes. For example, histone ubiquitination and methylation facilitate the recruitment of DNA repair factors to damaged sites, aiding in DNA repair and maintaining genome stability. |
| 5. Chromatin Remodeling |
Histone modifications are closely linked to chromatin structure and dynamics. They influence the folding and packaging of DNA into chromatin, regulating accessibility to DNA-binding proteins and controlling processes such as DNA replication, transcription, and DNA packaging into higher-order structures.
|
| 6. Cellular Senescence and Aging | Histone modifications have been implicated in cellular senescence, a state of stable cell cycle arrest that occurs in response to various stresses. Dysregulation of histone modifying enzymes can influence the onset and progression of cellular senescence, which is associated with aging and age-related diseases. |
| 7. Disease Pathogenesis | Dysregulation of histone modifying enzymes has been linked to various diseases, including cancer, neurological disorders, cardiovascular diseases, and autoimmune conditions. Altered histone modifications can contribute to aberrant gene expression patterns, disrupted chromatin structure, and impaired cellular processes, leading to disease development and progression. |
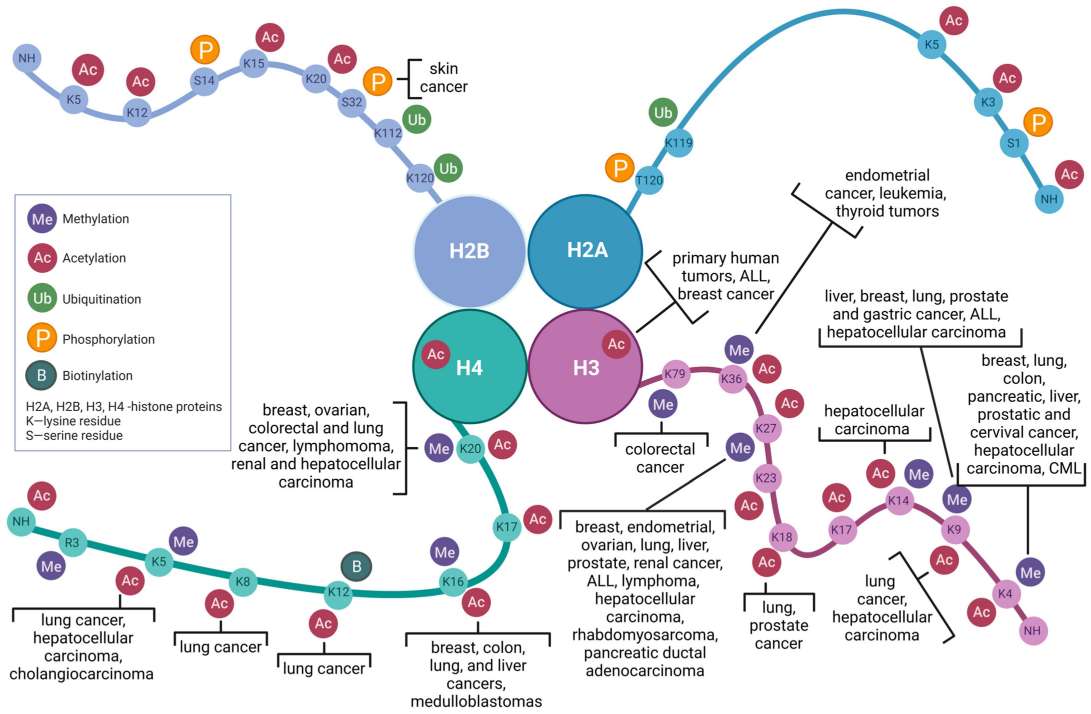 Fig.2 Correlation between histone modifications and distinct neoplastic entities. (Szczepanek J, et al., 2023)
Fig.2 Correlation between histone modifications and distinct neoplastic entities. (Szczepanek J, et al., 2023)Histone Modifying Enzymes in the Development of Diseases and Potential Therapeutic Targets
Histone modifying enzymes play a significant role in the development of various diseases, and their dysregulation can contribute to disease pathogenesis. Understanding the involvement of these enzymes in disease processes opens up opportunities for developing potential therapeutic targets. Here's an overview of their role in disease development and their potential as therapeutic targets:
Cancer
- Dysregulation of histone modifying enzymes is frequently observed in cancer. Altered histone acetylation, methylation, and other modifications can result in aberrant gene expression patterns that promote tumor growth and metastasis.
- Histone deacetylases (HDACs) are attractive therapeutic targets in cancer. HDAC inhibitors have been developed and approved for the treatment of certain cancers, as they can restore normal histone acetylation patterns, leading to transcriptional reprogramming and inhibition of cancer cell proliferation.
Neurological Disorders
- Histone modifications are implicated in neurological disorders such as Alzheimer's disease, Parkinson's disease, and Huntington's disease. Dysregulation of histone acetylation and methylation can affect gene expression in neurons, contributing to neurodegeneration and cognitive decline.
- Histone deacetylase inhibitors (HDACis) have shown promise in preclinical studies and clinical trials for neurodegenerative diseases. By modulating histone acetylation, HDACis can potentially restore neuronal function and ameliorate disease symptoms.
Cardiovascular Diseases
- Histone modifying enzymes have been implicated in cardiovascular diseases, including heart failure, atherosclerosis, and cardiac hypertrophy. Aberrant histone modifications can disrupt gene expression programs involved in cardiac remodeling and function.
- Targeting specific histone modifying enzymes, such as HDACs and histone methyltransferases, holds promise for treating cardiovascular diseases. Inhibitors of these enzymes have demonstrated potential in preclinical studies, showing beneficial effects on cardiac function and remodeling.
Autoimmune Conditions
- Histone modifications are involved in the dysregulation of immune responses seen in autoimmune conditions such as rheumatoid arthritis, lupus, and multiple sclerosis. Altered histone modifications can affect the expression of genes involved in immune regulation and inflammation.
- Modulating histone modifications through the use of inhibitors or activators of histone modifying enzymes represents a potential therapeutic strategy for autoimmune diseases. However, further research is needed to elucidate the specific roles of histone modifying enzymes in these conditions.
Epigenetic Therapies
- The dysregulation of histone modifying enzymes in various diseases has led to the development of epigenetic therapies. Small molecule inhibitors and activators targeting histone modifying enzymes, such as HDAC inhibitors and HAT activators, are being explored as potential treatments.
- Epigenetic therapies aim to restore normal histone modifications and gene expression patterns, providing a means to reprogram gene expression in disease states. Clinical trials are ongoing to assess the efficacy and safety of these therapies in different diseases.
While histone modifying enzymes hold promise as therapeutic targets, it is essential to carefully consider their specific roles and functions in each disease context. Further research is needed to fully understand the complexities of histone modifications and their therapeutic potential.
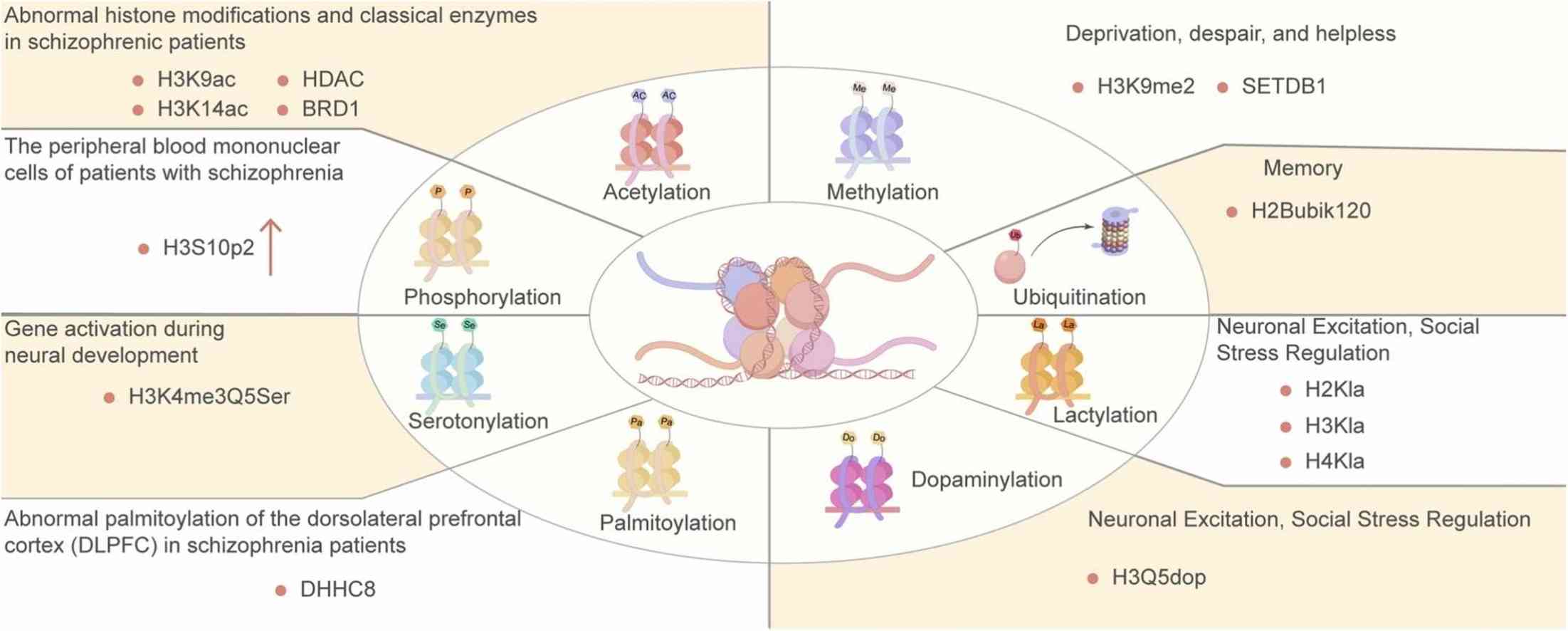 Fig.3 Abnormal histone modifications and classical enzymesin schizophrenic patients. (Chen YZ, et al., 2024)
Fig.3 Abnormal histone modifications and classical enzymesin schizophrenic patients. (Chen YZ, et al., 2024)Case Study
Case 1: Molina O, Vargiu G, Abad MA, et al. Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat Commun. 2016;7:13334.
The study highlights the findings of a study regarding the role of histone modifications, specifically H3K4me2 and H3K9ac, in centromere function and stability. Here is a summary of the key points:
1. Centromere composition: Centromeres are composed of specialized centrochromatin, which contains CENP-A nucleosomes and H3 nucleosomes carrying transcription-associated modifications.
2. 'In situ epistasis' analysis: The study employed an innovative approach called 'in situ epistasis' analysis. This involved targeting LSD2, a demethylase specific to H3K4me2, and synthetic modules with competing activities to a synthetic alphoidtetO HAC centromere. This approach allowed the separation of transcription from histone modifications at the centromere.
3. Role of H3K4me2: Loss of H3K4me2 was found to decrease centromeric transcription, CENP-A assembly, and stability. Additionally, it led to the spreading of H3K9me3 across the HAC, resulting in centromere inactivation.
4. Transcription and histone modifications: Surprisingly, transcription induced by CENP-28/Eaf6, associated with H4K12 acetylation, did not rescue the observed phenotype. In contrast, transcription induced by p65, associated with H3K9 acetylation, did rescue the phenotype. This suggests that mitotic transcription, along with histone modifications including H3K9ac, plays a crucial role in enabling CENP-A assembly and maintaining centrochromatin.
5. H3K4me2 and H3K9ac: The study indicates that H3K4me2 is required for transcription, while H3K9ac may act as a barrier to prevent heterochromatin spreading and inactivation of the kinetochore at human centromeres.
These findings highlight the significance of specific histone modifications and transcription in the regulation of centromere function and maintenance. H3K4me2 and H3K9ac emerge as critical players in the epigenetic landscape that enables CENP-A assembly and helps to maintain centromere stability.
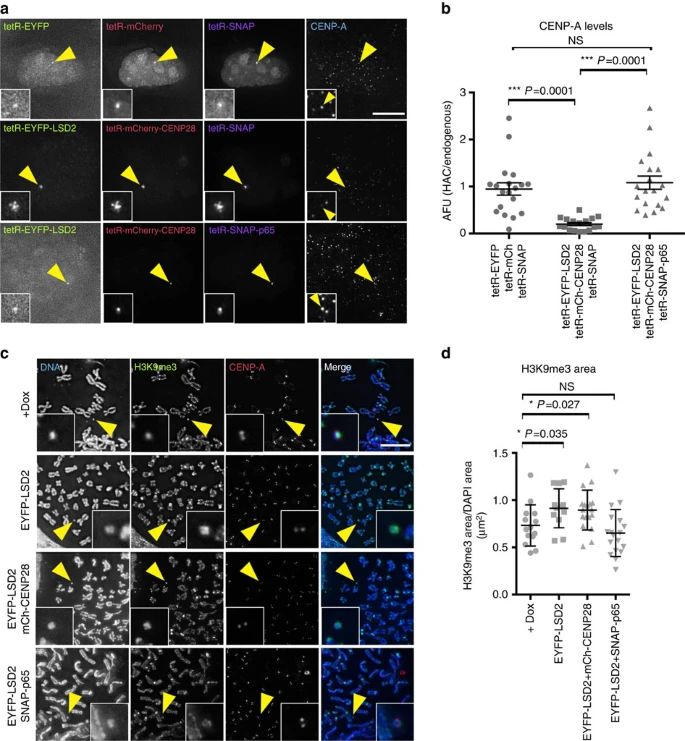 Fig.1 H3K4me2 and H3K9ac maintain the epigenetic signature of centrochromatin.
Fig.1 H3K4me2 and H3K9ac maintain the epigenetic signature of centrochromatin.Case 2: Stephens AD, Liu PZ, Banigan EJ, et al. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell. 2018;29(2):220-233.
The study discusses the influence of nuclear shape and architecture on various cellular processes and the association of abnormal nuclear morphology, specifically nuclear blebs, with several human conditions such as heart disease, aging, progeria, and cancer. The summary of the key findings is as follows:
1. Nuclear blebs and diagnostic markers: Nuclear blebs, abnormal protrusions in the nucleus, are considered diagnostic markers for various human afflictions. They are associated with alterations in both lamin proteins and chromatin.
2. Role of lamins and chromatin compaction: Previous studies have suggested that lamins play a role in determining nuclear morphology. However, the contributions of altered chromatin compaction in nuclear blebbing are not well understood.
3. Chromatin histone modification state and nuclear rigidity: The study demonstrates that the state of chromatin histone modifications influences nuclear rigidity. Modulating histone modifications is sufficient to induce or suppress nuclear blebbing. Treatment of mammalian cells with histone deacetylase inhibitors, which increase euchromatin, or histone methyltransferase inhibitors, which decrease heterochromatin, results in a softer nucleus and nuclear blebbing, without affecting lamins.
4. Histone demethylase inhibitors and nuclear blebbing: Conversely, treatment with histone demethylase inhibitors, which increase heterochromatin and chromatin nuclear rigidity, leads to reduced nuclear blebbing in lamin B1 null nuclei.
5. Rescue of nuclear morphology: Increased heterochromatin also rescues nuclear morphology in a model cell line for Hutchinson-Gilford progeria syndrome caused by mutant lamin A, as well as in cells from patients with the disease.
In summary, the histone modification state of chromatin is a significant factor in determining nuclear blebbing and morphology by influencing nuclear rigidity. While lamins play a role in nuclear morphology, altered chromatin compaction, driven by histone modifications, contributes to the development of nuclear blebs independently of lamins. Modulating chromatin histone modifications can affect nuclear rigidity and potentially impact cellular functions and diseases associated with abnormal nuclear morphology.
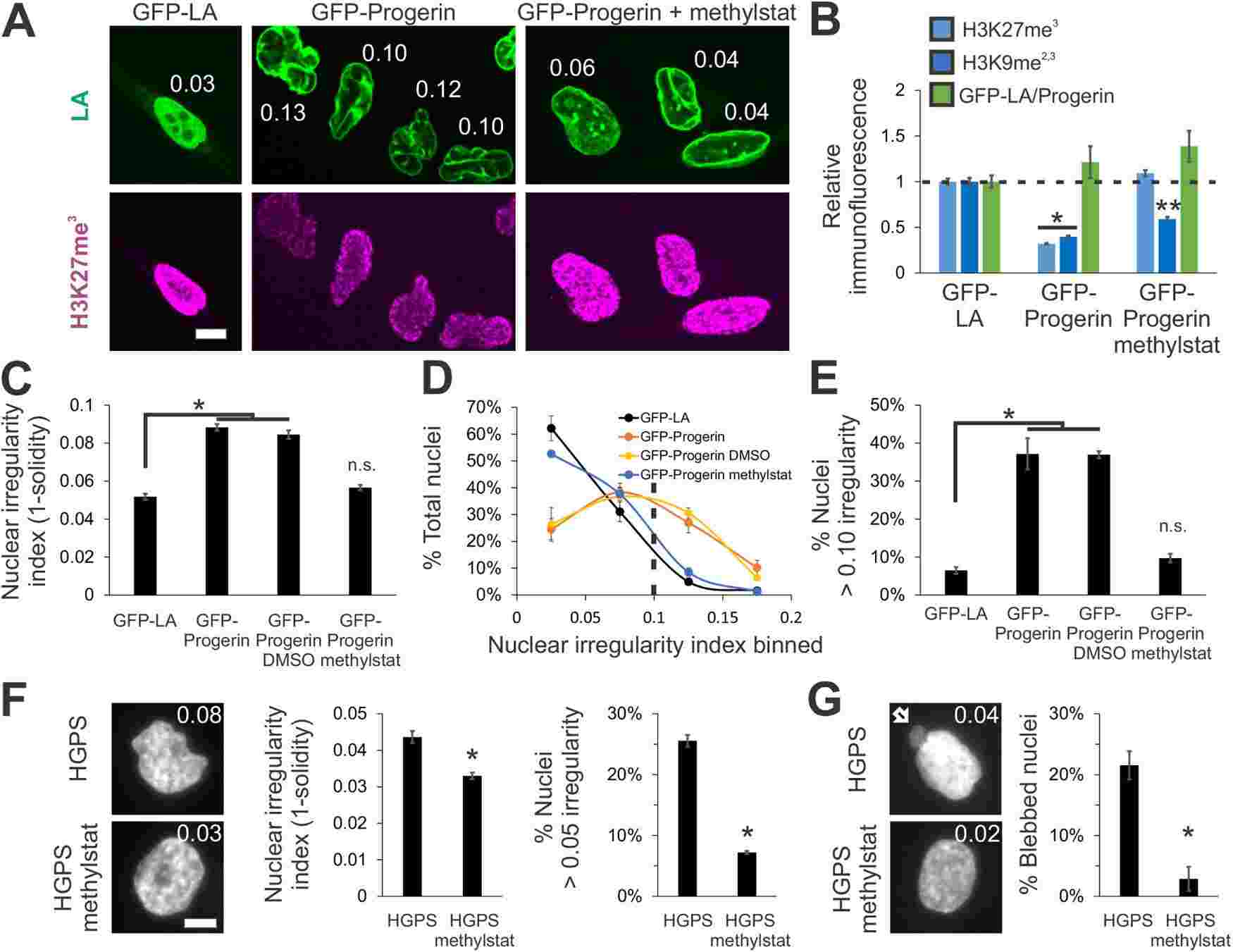 Fig.2 Increased heterochromatin via methylstat treatment rescues nuclear morphology in a model cell line and patient cells of laminopathy/aging disease HGPS caused by mutant lamin A.
Fig.2 Increased heterochromatin via methylstat treatment rescues nuclear morphology in a model cell line and patient cells of laminopathy/aging disease HGPS caused by mutant lamin A.Related References
- Graça, I., Pereira-Silva, E., Henrique, R. et al. Epigenetic modulators as therapeutic targets in prostate cancer. Clin Epigenet 8, 98 (2016).
- Chen YZ, Zhu XM, Lv P, et al. Association of histone modification with the development of schizophrenia. Biomed Pharmacother. 2024;175:116747.
- Szczepanek J, Tretyn A. MicroRNA-mediated regulation of histone-modifying enzymes in cancer: mechanisms and therapeutic implications. Biomolecules. 2023; 13(11):1590.

