SARS-CoV-2 Proteins and Their Target Proteins
Featured Interaction Proteins
| CAT.NO. | Sequence | PRODUCT NAME | IDENTITY | SPECIES | SOURCE |
|---|---|---|---|---|---|
| AXL-535H | Ala26-Pro440 | Recombinant Human AXL protein, His-tagged (Active) | AXL-His | Human | HEK293 |
| AXL-536H | Ala26-Pro440 | Recombinant Human AXL protein, Fc-tagged | AXL-Fc | Human | HEK293 |
| AXL-770H | Ala26-Pro449 | Recombinant Human AXL protein, His-tagged | AXL-His | Human | HEK293 |
| AXL-585H | Ala26-Pro449 | Recombinant Human AXL protein, His-Avi-tagged | AXL-His-Avi | Human | HEK293 |
| AXL-584HB | Ala26-Pro449 | Recombinant Human AXL protein, His-Avi-tagged, Biotinylated | AXL-His-Avi-Biotin | Human | HEK293 |
| AXL-249H | 473-end | Recombinant Human AXL Receptor Tyrosine Kinase, His-tagged, Active | AXL-His | Human | Sf 9 Insect cells |
| AXL-36H | 464-885(end) | Recombinant Human AXL protein, Flag-tagged, Biotinylated | AXL-Flag | Human | Insect cells |
| Cat.No. | Product Name | Identity | Species | Source |
|---|---|---|---|---|
| ACE2-736H | Recombinant Human ACE2, His-tagged | ACE2-His | Human | HEK293 |
| ACE2-68H | Recombinant Human ACE2, Fc-tagged | ACE2-hFc | Human | HEK293 |
| ACE2-185H | Recombinant Human ACE2, mouse IgG1 Fc-tagged | ACE2-mFc | Human | HEK293 |
| TMPRSS2-1856H | Recombinant Human TMPRSS2 Protein (106-492 aa), His-tagged | TMPRSS2-His | Human | Yeast |
| TMPRSS2-4899H | Recombinant Human TMPRSS2, his-tagged | TMPRSS2-His | Human | HEK293 |
| ACE2-229H | Recombinant Human ACE2 protein, His/Avi-tagged | ACE2-His/Avi | Human | Mammalian cells |
Druggable Targets
Coronavirus Proteins
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-588S | Recombinant SARS-CoV-2 Nucleocapsid (S202N) Protein, His-tagged | E.coli | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-479S | Recombinant SARS-CoV-2 (2019-nCoV) Nucleocapsid(P13L) Protein, His-tagged | E.coli | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| S-425S | Recombinant SARS-CoV-2 (2019-nCoV) Spike RBD (N501Y) Protein, Fc-tagged | HEK293 | Fc |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-587S | Recombinant SARS-CoV-2 Nucleocapsid (P67S, P199L) Protein, His-tagged | E.coli | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-405S | Recombinant SARS-CoV-2 (2019-nCoV) Nucleocapsid (D377Y) Protein, His-tagged | E.coli | His |
| N-406S | Recombinant SARS-CoV-2 Nucleocapsid (R203M, D377Y) Protein, His-tagged | E.coli | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-408S | Recombinant SARS-CoV-2 Nucleocapsid (P67S, R203M, D377Y) Protein, His-tagged | E.coli | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-589S | Recombinant SARS-CoV-2 Nucleocapsid (G18S, A119S, A217S, M234I, E367Q) Protein, His-tagged | E.coli | His |
| S-483S | Recombinant SARS-CoV-2 Spike S1+S2 ECD (YH145-146 deletion, E484K, D614G) Protein, His-tagged | Insect Cell | His |
| S-490S | Recombinant SARS-CoV-2 Spike S1 (H49Y, YH145-146 deletion, E484K, D614G) Protein, His-tagged | HEK293 | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| S-423S | Recombinant SARS-CoV-2 (2019-nCoV) Spike S1 (W152C, L452R, D614G) Protein, His-tagged | HEK293 | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-585S | Recombinant SARS-CoV-2 Nucleocapsid (A12G, T205I) Protein, His-tagged | E.coli | His |
| S-489S | Recombinant SARS-CoV-2 (2019-nCoV) Spike S1 (Q677H) Protein, His-tagged | HEK293 | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| S-417S | Recombinant SARS-CoV-2 Spike S1+S2 ECD (T95I, G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H) Protein, His-tagged | Insect Cell | His |
| S-419S | Recombinant SARS-CoV-2 Spike S1 (T95I, G142D, E154K, L452R, E484Q, D614G, P681R) Protein, His-tagged | HEK293 | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-584S | Recombinant SARS-CoV-2 Nucleocapsid (P199L, M234I) Protein, His-tagged | E.coli | His |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| S-474S | Recombinant SARS-CoV-2 (2019-nCoV) Spike S1(HV69-70 deletion, Y453F, D614G) Protein, His-tagged | HEK293 | His |
| S-475S | Recombinant SARS-CoV-2 (2019-nCoV) Spike RBD(Y453F) Protein, His-tagged | HEK293 | His |
| S-476S | Recombinant SARS-CoV-2 (2019-nCoV) Spike RBD (Y453F) Protein, rFc-tagged | HEK293 | rFc |
| Cat.No. | Product Name | Source | Tag |
|---|---|---|---|
| N-586S | Recombinant SARS-CoV-2 Nucleocapsid (A119S, R203K, G204R, M234I) Protein, His-tagged | E.coli | His |
Backgound
The COVID-19 outbreak has caused large numbers of death, as well as social and economic chaos worldwide. SARS-CoV-2 virus is the causative agent of this respiratory disease.
The scientific community has known that SARS-CoV-2 binds to host cell receptors through its surface spike (S) proteins and is endocytosed into cells through clathrin-mediated membrane fusion. The relationship and affinity between the host cell receptor and the virus surface protein determine the infectivity and cytophilic characteristics of the virus. The pathogenicity and invading of the virus are related to the type and function of the host cell. Crystal structure analysis confirmed that angiotensin-converting enzyme 2 (ACE2) is the receptor for SARS-CoV-2 S protein, and the affinity of the two is 10-20 times that of SARS-CoV and ACE2, which is the reason of SARS-CoV-2 super infectivity.
So far, no clinically available antiviral drugs have been developed for SARS-CoV, SARS-CoV-2, or MERS-CoV. Some studies used affinity purification-mass spectrometry analysis that have identified hundreds of protein interactions between SARS-CoV-2 proteins and human proteins, including DNA replication (Nsp1), epigenetic and gene expression regulators (Nsp5, Nsp8, Nsp13, E), vesicle trafficking (Nsp6, Nsp7, Nsp10, Nsp13, Nsp15, Orf3a, E, Orf8), lipid modification (Spike), RNA processing and regulation (Nsp8, N), ubiquitin ligases (Orf10), signaling (Nsp8, Nsp13, N, Orf9b), nuclear transport machinery (Nsp9, Nsp15, Orf6), cytoskeleton (Nsp1, Nsp13), mitochondria (Nsp4, Nsp8, Orf9c), and extracellular matrix (Nsp9). Results from data analysis suggested that the viral proteins connect to a wide array of biological processes, including protein trafficking, translation, transcription, and ubiquitination regulation.
A total of 332 high confidence SARS-CoV-2-human protein-protein interactions (PPIs) have been identified. Among these, using a combination of a systematic chemoinformatic drug search with a pathway centric analysis, researchers have uncovered 66 druggable human proteins or host factors targeted by close to 70 different drugs and compounds, including FD A approved drugs, compounds in clinical trials as well as preclinical compounds. Additionally, this proteomic/chemoinformatic analysis gives these potential therapeutic perturbations a mechanistic context.
Creative BioMart has collected many SARS-CoV-2 related proteins and druggable target proteins, which can help you discover antiviral therapies against SARS-CoV-2 and other deadly coronavirus strains.
B.1.1.529 contains at least 50 mutations, 30 of which are on the spike protein (S protein). Not surprisingly, spike protein has become the research and development target of most vaccines. Some of the mutations are known to affect infectivity and immune escape, while others are newly identified. It is currently unknown what will happen when these mutations combine and their effects on the Sars-CoV-2 vaccine.
Coronavirus uses its spike glycoprotein (S), a main target for neutralization antibody, to bind to its receptor, and mediate membrane fusion and virus entry. Its receptor binding domain (RBD) has a high affinity with the ACE2 receptor protein on the surface of human cells. The S protein is an ideal entry point for vaccine design. The neutralizing antibody binds to the viral S protein RBD to competitively block the virus binding to the ACE2 protein and infect cells, achieving the effect of immune prevention and even treatment.
Creative BioMart is still committed to overcoming the challenges posed by the COVID-19 pandemic. We have just added a new panel of mutant RBD/spike and nucleocapsid proteins. In addition to the Omicron variant, Creative BioMart also provides other COVID variants (Alpha, Beta, Gamma, Delta, Eta, Iota, Kappa, and Lambda). We will continue to update our catalog to include new mutant proteins.
We can now offer mutant spike RBD proteins, which are thermally stable and can produce antibodies with neutralizing activity. We will continue to work together with the worldwide scientists to find the path forward for today’s challenges, and to support tomorrow’s breakthroughs.
South Africa has detected a new mutant of the Sars-CoV-2-B.1.1.529. The World Health Organization (WHO) included B.1.1.529 on the Variant of Concern (VOC), named Omicron.
Advantages
Consistency and Safety: Recombinant proteins can be produced consistently and in high quantities, ensuring they exhibit the same characteristics time and again. They do not carry the risk of infection as they are not part of the live virus.
Easy Scalability: They can be easily scaled up for mass production, making them suitable for large-scale screening, vaccine development, etc.
Controlled study: They allow for the study of individual viral proteins in isolation, which is not possible with the complete virus.
Affordability: It's not overly expensive to produce recombinant proteins, which is an advantage, especially in a public health crisis where cost-effectiveness is paramount.
Manipulation Capability: Recombinant proteins can be manipulated and modified to study different aspects of the virus, such as how mutations could potentially affect its function and behavior.
Applications
SARS-CoV-2 strain mutant proteins and virus interaction proteins have several applications related to research, diagnostics, and therapeutics for COVID-19.
- Study of Virus-Host Interactions: It is very crucial for understanding how the virus infects human cells. By examining the interaction between the SARS-CoV-2 proteins and human cells, researchers can gain valuable insights into the life cycle of the virus and how it exploits host cells.
- Drug Development: The SARS-CoV-2 strain mutant proteins and virus interaction proteins have been extensively used in the development of potential anti-viral drugs. Understanding the structure and function of these proteins can be used to design drugs that can potentially inhibit these proteins, preventing the virus from replicating.
- Vaccine Development: The spike protein of SARS-CoV-2, which is important for the virus's ability to infect human cells, is the target for most COVID-19 vaccines. Understanding how this protein and its variants interact with human cells helps in the development of more effective vaccines.
- Diagnostics: SARS-CoV-2 strain mutant proteins can be used in the development of diagnostic tools. For instance, tests that detect the presence of viral proteins in the blood or other fluids can be used to confirm COVID-19 infection.
- Therapeutics: Antibodies designed to target specific viral proteins can act as therapeutic drugs. For example, monoclonal antibody treatments for COVID-19 work by binding to the spike protein and blocking the virus's ability to attach to and infect cells.
- Understanding Virus Evolution: Studying the changes and variations in SARS-CoV-2 strain mutant proteins over time can help understand how the virus evolves, giving critical insights for public health responses and future vaccine development.
- Epidemiological Surveillance: The study of different strains can be used to trace the spread and mutation of the virus in the population, providing valuable information for predicting future outbreaks and monitoring the progress of the pandemic.
Case Study
Case 1: Stein SR, Ramelli SC, Grazioli A, et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature. 2022 Dec;612(7941):758-763. doi: 10.1038/s41586-022-05542-y. Epub 2022 Dec 14. PMID: 36517603; PMCID: PMC9749650.
The Researchers carried out complete autopsies on 44 patients who died with COVID-19, with extensive sampling of the central nervous system in 11 of these patients, to map and quantify the distribution, replication and cell-type specificity of SARS-CoV-2 across the human body, including the brain, from acute infection to more than seven months following symptom onset. We show that SARS-CoV-2 is widely distributed, predominantly among patients who died with severe COVID-19, and that virus replication is present in multiple respiratory and non-respiratory tissues, including the brain, early in infection.
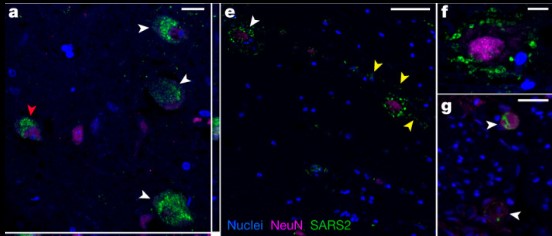
Fig1. SARS-CoV-2 protein and RNA expression in human CNS tissues. a, High-magnification visualization of hypothalamus from P38 labelled for SARS-CoV-2 N protein (green) and neuronal nuclear (NeuN) protein (magenta), demonstrating viral-specific protein expression in neurons (white arrowheads) by IF. The z-stack orthogonal views to the right and bottom of a demonstrate NeuN labelling in the nucleus and SARS-CoV-2 protein in the cytoplasm of the cell (red arrowhead).
Case 2: Ju X, Zhu Y, Wang Y, Li J, et al. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLoS Pathog. 2021 Mar 12;17(3):e1009439. doi: 10.1371/journal.ppat.1009439. PMID: 33711082; PMCID: PMC7990224.
ARS-CoV-2 is classified as a biosafety level-3 (BSL-3) agent, impeding the basic research into its biology and the development of effective antivirals. The authors developed a biosafety level-2 (BSL-2) cell culture system for production of transcription and replication-competent SARS-CoV-2 virus-like-particles (trVLP). This trVLP expresses a reporter gene (GFP) replacing viral nucleocapsid gene (N), which is required for viral genome packaging and virion assembly (SARS-CoV-2 GFP/ΔN trVLP).
They developed a convenient and efficient SARS-CoV-2 reverse genetics tool to dissect the virus life cycle under a BSL-2 condition. This powerful tool should accelerate our understanding of SARS-CoV-2 biology and its antiviral development.
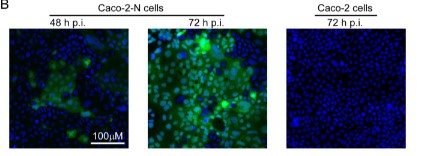
Fig1. The recombinant SARS-CoV-2-GFP/ΔN trVLP can propagate with the help of viral N protein. GFP expression was observed in Caco-2 or Caco-2-N cells using microscopy at indicated time point after inoculation; Representative images from one of three independent experiments.
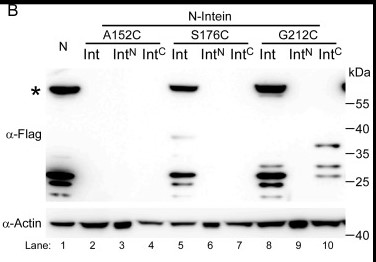
Fig2. Reconstitution of functional N protein by intein-mediated protein splicing. Western blot (WB) analysis of lysates from Caco-2 cells transduced with either full-length N or intein-N lentiviruses. The star indicates the full-length N protein. The WB is representative of three independent experiments.
Case 3: Fink K, Nitsche A, Neumann M, Grossegesse M, Eisele KH, Danysz W. Amantadine Inhibits SARS-CoV-2 In Vitro. Viruses. 2021 Mar 24;13(4):539. doi: 10.3390/v13040539. PMID: 33804989; PMCID: PMC8063946.
Even if vaccines are approved, the need for antiviral agents against SARS-CoV-2 will persist. However, unequivocal evidence for efficacy against SARS-CoV-2 has not been demonstrated for any of the repurposed antiviral drugs so far. Amantadine was approved as an antiviral drug against influenza A, and antiviral activity against SARS-CoV-2 has been reasoned by analogy but without data. The authors tested the efficacy of amantadine in vitro in Vero E6 cells infected with SARS-CoV-2.
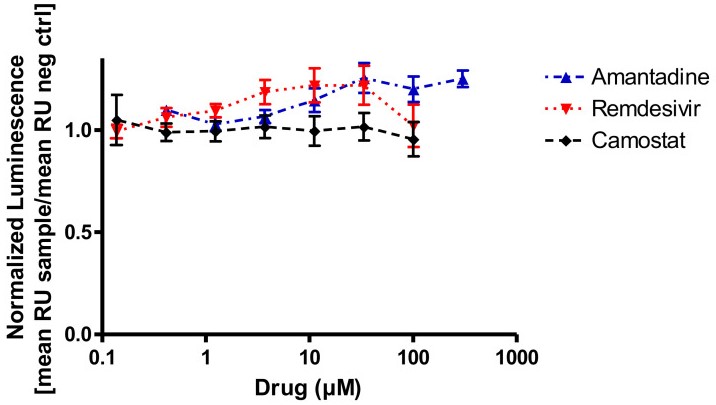
Fig1. In vitro binding interference of the SARS-CoV-2 spike protein to recombinant ACE2 and amantadine, remdesivir, or camostat mesylate. None of the drugs inhibit SARS-CoV-2 binding to its target protein
Related Resources
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions