Protein SUMOylation Assay
Creative BioMart provides a comprehensive Protein SUMOylation Assay Service designed to facilitate the in vitro study of SUMOylation and its impact on protein function. Using our optimized assay system, we generate SUMOylated proteins by covalently attaching SUMO-1, SUMO-2, or SUMO-3 peptide substrates to specific lysine residues of target proteins via isopeptide bonds. Our service offers robust and reproducible results suitable for applications ranging from basic research to therapeutic discovery. With customizable assay formats, advanced detection methods, and expert scientific support, Creative BioMart ensures accurate and high-quality data that meet the needs of both academic researchers and pharmaceutical developers.
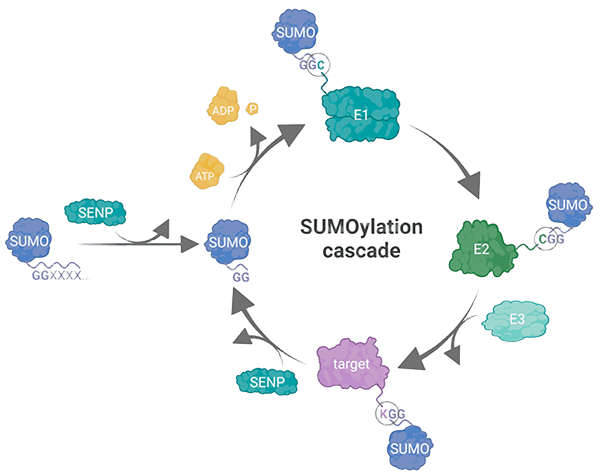
Understanding Protein SUMOylation and Its Biological Role
Protein SUMOylation is a post-translational modification (PTM) in which Small Ubiquitin-like Modifier (SUMO) proteins are covalently attached to target proteins. This reversible modification plays a critical role in diverse cellular processes such as transcriptional regulation, DNA repair, nuclear transport, and signal transduction. Dysregulation of SUMOylation has been implicated in cancers, neurodegenerative disorders, and other diseases, making it a valuable target for biomedical research and drug development. By providing reliable and efficient SUMOylation assays, Creative BioMart supports researchers in understanding the mechanistic roles of SUMOylation and exploring novel therapeutic interventions.
Protein SUMOylation Assay Services
Our Protein SUMOylation Assay Service includes:
-
In vitro SUMOylation assay systems with SUMO-1, SUMO-2, or SUMO-3 peptide substrates.
-
Covalent linkage of SUMO peptides to target proteins at lysine residues through isopeptide bonds.
-
Western blot detection and analysis to confirm SUMOylation, using optimized antibody conditions.
-
Custom assay development to study specific proteins of interest or explore SUMOylation in mutant versus wild-type contexts.
-
Functional assessment of SUMOylated proteins to investigate their roles in transcription, protein stability, or pathways.
Service Workflow

Applications
- Investigating SUMOylation and transcription factor activity
- Exploring SUMOylation in cellular regulation
- Identifying novel SUMO-modified proteins
Representative Results
To demonstrate the specificity of our SUMOylation analysis service, we performed a Western blot assay on p53. This result highlights our ability to distinguish between wild-type and mutant SUMOylation reactions while confirming the absence of nonspecific labeling in control experiments.
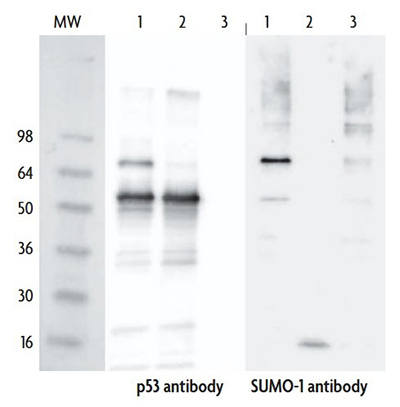
Figure 1. Western blot analysis of in vitro SUMOylation of p53 protein. p53 was incubated with either wild-type or mutated isoforms of SUMO-1 and probed with anti-p53 (1:5000) and anti–SUMO-1 (1:4000) antibodies. SUMOylation of p53 was detected only in the wild-type SUMO-1 conjugation reaction (Lane 1). No modification was observed with the mutated SUMO-1 isoform (Lane 2) or in the control reaction lacking p53 (Lane 3).
Why Partner with Us for SUMOylation Assays
- Comprehensive Expertise: Our team specializes in PTMs and provides expert consultation for SUMOylation studies.
- Customizable Assays: Flexible assay design to fit specific proteins, isoforms, or experimental conditions.
- High Sensitivity & Specificity: Optimized protocols ensure accurate detection with minimal background.
- Diverse Applications: From basic research to drug discovery, our assays support multiple scientific needs.
- Reliable Quality Control: ISO13485-based processes ensure reproducibility and consistency across projects.
- End-to-End Support: From experimental design to data interpretation, we provide complete project guidance.
Applications and Case Studies
Case 1: In vitro SUMOylation assay to study SUMO E3 ligase activity
Yang et al., 2018. doi:10.3791/56629
SUMOylation is carried out through a sequential enzyme cascade involving E1-activating (SAE1/SAE2), E2-conjugating (Ubc9), and, optionally, E3-ligase enzymes (e.g., PIAS family, RanBP2, Pc2), with E3 ligases enhancing specificity and efficiency. While SUMOylated proteins can be detected in vivo via immunoprecipitation and immunoblotting, assessing E3 ligase activity requires in vitro reconstitution with purified components. A modified protocol allows consistent detection of SUMO conjugation, exemplified by viral K-bZIP E3 ligase activity on p53, enabling identification of novel SUMO E3 ligases and their paralog specificity.
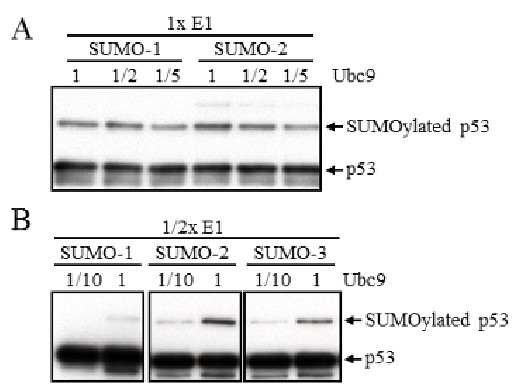
Figure 2. Determination of the minimal amount of SUMO enzymes used in the in vitro SUMOylation assay. (A) p53 SUMOylation was evaluated in an in vitro SUMOylation assay that included one half and one fifth the amount of Ubc9 enzyme used in standard protocol. After 3 hours of in vitro SUMOylation reaction, SUMOylated p53 enzymes were examined by Western blot with anti-p53 antibody. (B) In vitro SUMOylation of p53 was further analyzed by using half the amount of E1 enzyme in combination with one-tenth the amount of Ubc9 used in the standard protocol. (Yang et al., 2018)
Case 2: Development of a high-throughput screen to detect inhibitors of TRPS1 SUMOylation
Brandt et al., 2013. doi:10.1089/adt.2012.501
Small ubiquitin-like modifier (SUMO) proteins are reversibly conjugated to specific lysines on target proteins, but quantitative assay formats for large-scale screening have been limited. To identify inhibitors of Ubc9-dependent SUMOylation, a high-throughput fluorescence polarization assay was developed using a minimal hexapeptide substrate, TMR-VVK1201TEK, representing the primary SUMOylation site of TRPS1. This format enabled screening of a large compound library, confirming 728 hits. While no specific noncovalent Ubc9 inhibitors were identified, several diaminopyrimidine compounds were found to inhibit SUMOylation with IC50 values of 12.5 μM and acted as competitive substrates, demonstrating the assay’s utility for identifying modulators of SUMO conjugation.
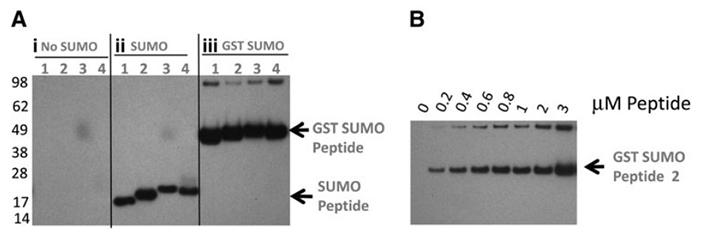
Figure 3. Gel-based demonstration of E3-independent SUMOylation of peptide substrates by the coupled E1:Ubc9 assay format. Synthetic N-acetyl biotin peptide substrates based on known SUMOylation sites in p53 (K356), TRPS1 (K1192 and K1201), and TCF4 (K297) were evaluated in an in vitro SUMOylation assay. (A) Detection of SUMO-1 or GST-SUMO-1 peptide adducts by Western blotting and detection of biotinylated peptides using HRP-SA. Peptide substrate sequences are indicated in Table 1. (i) No SUMO-1 addition; (ii) SUMO-1 addition; (iii) GST-SUMO-1 addition. (B) Conjugation of peptide 2 with GST-SUMO-1 (4.2 mM) over the range of peptide concentrations indicated. (Brandt et al., 2013)
Researcher Feedback on Our SUMOylation Assay Services
"Creative BioMart’s Protein SUMOylation Assay played a crucial role in our transcription factor research project. Their team helped us evaluate how SUMOylation impacted p53 activity, providing high-quality Western blot data that were reproducible and publication-ready. The level of technical detail in their data package allowed us to rapidly move forward with functional studies. Their responsiveness and scientific expertise made the collaboration seamless."
— Principal Investigator | Academic Research Institute
"We engaged Creative BioMart to identify novel SUMOylation targets in a panel of signaling proteins relevant to cancer biology. Their systematic workflow, from in vitro SUMOylation reactions to downstream detection, gave us clear and convincing results. Not only did they deliver on time, but their ability to troubleshoot complex protein modifications exceeded our expectations. The insights we gained have already fed into our drug discovery pipeline."
— Senior Scientist, Oncology Research | Mid-Sized Biotech Company
"As part of our neurodegeneration program, we needed reliable data on the role of SUMOylation in stress response pathways. Creative BioMart provided a customized SUMOylation assay service that integrated both SUMO-1 and SUMO-2 substrates, helping us compare isoform-specific effects. The clarity of their experimental design and their thorough interpretation of results saved us months of internal development time."
— Director of Neuroscience R&D | Global Pharmaceutical Company
"Our team collaborated with Creative BioMart to assess SUMOylation-mediated regulation of DNA repair proteins. Their experts guided us in selecting appropriate assay conditions and validated the SUMOylation status with high sensitivity. The data package was detailed, well-annotated, and suitable for both internal reporting and future regulatory submissions. Their professionalism and technical know-how make them a trusted partner in our preclinical research efforts."
— Head of Translational Biology | International Biopharma Company
FAQs About Protein SUMOylation Assay Services
-
Q: What types of proteins can be analyzed with your SUMOylation assay?
A: Our assay supports a wide range of target proteins, including transcription factors, signaling proteins, and regulatory enzymes. We provide carboxy-terminal SUMO-1, SUMO-2, or SUMO-3 substrates that covalently modify specific lysine residues, enabling detailed functional studies across diverse protein classes. -
Q: Can I analyze SUMOylation effects for mutated or isoform-specific proteins?
A: Yes. We can examine wild-type as well as mutated isoforms of proteins to assess SUMOylation patterns, functional consequences, and isoform-specific regulation using high-sensitivity detection methods like Western blotting. -
Q: Do you provide customized assay designs?
A: Absolutely. We tailor assay conditions—including substrate selection, protein concentration, and reaction buffers—to meet your specific research needs. This flexibility ensures accurate evaluation of SUMOylation under physiological or experimental conditions relevant to your project. -
Q: What detection methods are used to measure SUMOylation?
A: We employ highly sensitive detection techniques such as Western blotting and immunodetection using SUMO-specific antibodies. Our team ensures clear and quantitative results for both in vitro SUMOylation and comparative studies between multiple protein isoforms. -
Q: How much protein sample is required for the assay?
A: Our optimized assay can work with microgram-level protein quantities. For most studies, standard recombinant protein preparations are sufficient, and our team can advise on minimum requirements for specific experimental designs. -
Q: Can this service help identify novel SUMOylation targets?
A: Yes. We offer systematic screening workflows to identify previously unknown SUMOylated proteins, enabling insights into regulatory networks and new avenues for therapeutic research. -
Q: What is included in your service package?
A: Our service includes assay setup, SUMOylation reactions, detection, data collection, and a detailed report. We provide all necessary reagents, technical support, and interpretation guidance, ensuring reproducible and publication-ready results. -
Q: How quickly can results be delivered?
A: Typical turnaround times are project-dependent, but our streamlined in vitro workflow allows rapid generation of reliable SUMOylation data, often within 2–4 weeks from sample submission.
Resources
Related Services
- Transcription Factor Assay
- Enzyme Activity Assay
- SUMO-tag Protein Production
- Protein Characterization
Related Products
References:
- Brandt M, Szewczuk LM, Zhang H, et al. Development of a high-throughput screen to detect inhibitors of TRPS1 SUMOylation. ASSAY and Drug Development Technologies. 2013;11(5):308-325. doi:10.1089/adt.2012.501
- Yang WS, Campbell M, Kung HJ, Chang PC. In vitro SUMOylation assay to study SUMO E3 ligase activity. JoVE. 2018;(131):56629. doi:10.3791/56629
- Wild N, Kaiser CS, Wunderlich G, Liebau E, Wrenger C. Protein SUMOylation and its functional role in nuclear receptor control. Receptors. 2024;3(3):408-424. doi:10.3390/receptors3030020
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

