Protein Phosphorylation Site Identification and Mapping Service
Creative BioMart offers a comprehensive Protein Phosphorylation Site Identification and Mapping Service for in-depth characterization of phosphorylated proteins. Our platform allows detection of phosphosites in proteins phosphorylated in vivo and purified by immunoprecipitation, or in recombinant proteins phosphorylated in vitro using purified kinases. Utilizing advanced techniques such as collision-induced dissociation (CID), electron-transfer dissociation (ETD), and radiolabelled 32P analyses coupled with HPLC and mass spectrometry, we provide high-resolution mapping of phosphorylation sites. This service enables researchers to uncover key post-translational modifications critical for protein function, signaling pathways, and drug target validation.
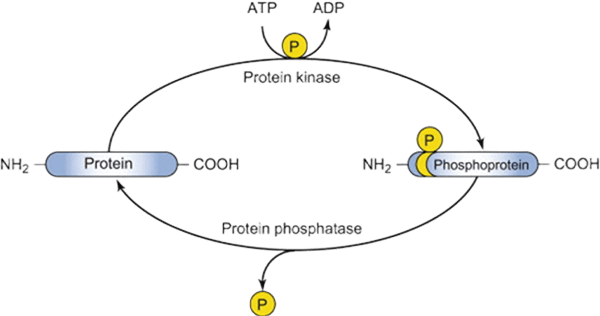
Background: Importance of Protein Phosphorylation Site Identification and Mapping
Protein phosphorylation is a crucial post-translational modification regulating numerous cellular processes, including signal transduction, enzyme activity, and protein-protein interactions. Identifying the exact phosphorylation sites is vital for understanding molecular mechanisms, elucidating cellular pathways, and developing therapeutic strategies. Traditional methods often lack the sensitivity or resolution to detect low-abundance phosphorylation events, while advanced mass spectrometry and radiolabeling approaches enable precise mapping and quantification. Creative BioMart’s expertise in these technologies provides researchers with reliable, high-resolution phosphorylation site identification for both in vivo and in vitro phosphorylated proteins.
Figure 1. An example of protein with phosphorylation site. (Johnson et al., 2015)
Protein Phosphorylation Site Identification and Mapping Services
Our Phosphorylation Mapping Capabilities
-
Comprehensive Sample Support
Analyze both in vivo immunoprecipitated proteins and in vitro kinase-phosphorylated recombinant proteins for maximum flexibility.
-
Advanced Mass Spectrometry
Use CID and ETD techniques for precise identification of phosphorylation sites, including labile phospho-groups.
-
Radiolabelled 32P Analysis
Detect low-abundance phosphorylation events with HPLC separation and on-line radioactivity detection.
-
Phosphosite Identification
Combine mass spectrometry and, if needed, solid-phase Edman degradation to confirm site-specific phosphorylation.
-
Detailed Data Reporting
Receive publication-ready results including annotated spectra, peptide mapping, phosphosite localization, stoichiometry, and potential kinase-substrate relationships.
-
Versatile Workflow Integration
Customizable analysis that fits diverse experimental designs, from basic research to drug discovery applications.
Service Workflow

Our Advantages in Phosphorylation Site Analysis
- World-Class Expertise: Recognized globally for pioneering phosphorylation site identification techniques.
- Comprehensive Methodology: Combines mass spectrometry, radiolabeling, and Edman degradation for high-confidence site mapping.
- High Sensitivity: Detects low-abundance phosphosites undetectable by conventional methods.
- Versatile Sample Types: Compatible with in vivo immunoprecipitated proteins and in vitro kinase substrates.
- Publication-Ready Data: Results formatted for direct inclusion in manuscripts and reports.
- Customized Analysis: Tailored workflows to accommodate unique experimental designs or specialized phosphorylation studies.
Case Studies in Protein Phosphorylation Site Mapping
Case 1: High-throughput quantification of adenylate turnover in marine microbes
Lanpher and Popendorf, 2022. doi:10.1002/lom3.10499
A high-throughput method was developed to quantify ATP, ADP, and AMP turnover rates in marine microbes by simultaneously measuring intracellular concentrations and phosphorylation rates. Microbial adenylates were extracted using boiling 20 mM Tris buffer, purified, and analyzed via optimized high-performance liquid chromatography. Radiolabeled phosphate (32P i ) incubations enabled determination of phosphorus uptake and phosphorylation rates. This approach allows direct assessment of microbial growth rates, energy charge, metabolic turnover, and adenylate storage. Validation with Biscayne Bay samples revealed turnover times of 12, 15, and 73 minutes for ATP, ADP, and AMP, respectively, providing a powerful tool for monitoring microbial metabolism across marine environments.
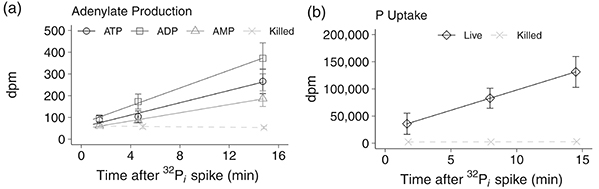
Figure 2. The linear incorporation of 32P-radiolabeled phosphate (32P i ) spikes into (a) adenosine triphosphate (ATP, circles), adenosine diphosphate (ADP, squares), and adenosine monophosphate (AMP, triangles) in a representative sample from Biscayne Bay seawater. (b) Total 32P i uptake by the microbial community in the Biscayne Bay seawater (diamonds). Control seawater killed with paraformaldehyde (0.5% final concentration) was measured for both protocols and is on both plots (killed; X dashed lines). (Lanpher and Popendorf, 2022)
Case 2: HIPK2-HDAC3-p65 module as a therapeutic target in inflammation
Zhang et al., 2021. doi:10.1073/pnas.2021798118
Uncontrolled inflammation contributes to diseases such as cancer and sepsis, yet therapies targeting specific cytokines have had limited success. This study identifies homeodomain-interacting protein kinase 2 (HIPK2) as a nuclear regulator that restrains NF-κB activation in macrophages. HIPK2 phosphorylates histone deacetylase 3 (HDAC3) at serine 374, inhibiting its activity, reducing p65 deacetylation, and thereby suppressing inflammatory responses. HIPK2 deficiency exacerbates LPS-induced endotoxemia, sepsis, and colorectal cancer in mice. Targeting the HIPK2-HDAC3-p65 axis offers a potential therapeutic strategy to control excessive inflammation, providing new avenues for treating inflammation-related diseases.
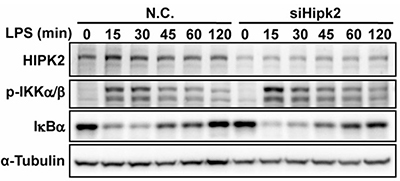
Figure 3. The phosphorylation levels of IKKα/β and the degradation of IκB were measured in LPS-stimulated control and Hipk2 knockdown PEMs. (Zhang et al., 2021)
Protein Phosphorylation Site Identification Client Reviews
“The Creative BioMart team provided invaluable support in mapping phosphorylation sites on our kinase of interest. Their expertise in CID and ETD mass spectrometry allowed us to identify novel regulatory sites that were critical for downstream functional studies. The detailed reports and data interpretation helped us accelerate our signaling pathway research significantly.”
— Senior Scientist | Biotech Research Institute
“We collaborated with Creative BioMart to analyze phosphorylation dynamics in a cell cycle-related protein. Their 32 P-labelling and HPLC-based approach offered unmatched sensitivity, revealing low-abundance phosphosites that standard methods missed. The results were instrumental in designing our follow-up functional assays.”
— R&D Director | Pharmaceutical Company
“For our drug discovery project targeting MAPK signaling, Creative BioMart’s phosphorylation mapping service was exceptional. They not only identified key phosphorylation sites but also provided clear mechanistic insights, helping us refine our inhibitor design strategy.”
— Principal Investigator | Academic Medical Center
“Creative BioMart’s mapping of phosphorylation sites on a recombinant transcription factor exceeded our expectations. Their workflow was efficient, and the technical team was responsive to our project-specific questions. The high-quality data greatly streamlined our publication and patent preparation process.”
— Head of Protein Chemistry | Global Biopharma Company
FAQs About Protein Phosphorylation Site Identification and Mapping
-
Q: What types of proteins can you analyze for phosphorylation sites?
A: We can analyze both in vivo phosphorylated proteins purified by immunoprecipitation and recombinant proteins phosphorylated in vitro using purified kinases. This flexibility allows us to handle diverse research targets, from signaling proteins to transcription factors. -
Q: Which methods do you use for phosphorylation site identification?
A: Our team employs advanced mass spectrometry techniques including Collision-Induced Dissociation (CID) and Electron-Transfer Dissociation (ETD), as well as 32P-labelled site analysis with HPLC separation. These approaches ensure high sensitivity and accurate site mapping. -
Q: Can you detect low-abundance or hard-to-find phosphorylation sites?
A: Yes. Our combination of radiolabelled detection, off-line HPLC separation, and mass spectrometry allows us to identify low-abundance or substoichiometric phosphorylation sites that conventional methods often miss. -
Q: How do you report the results?
A: We provide comprehensive, easy-to-interpret reports that include phosphorylation site locations, spectra, and validation data. This format supports downstream functional studies, publication, or patent applications. -
Q: How customizable is the service?
A: We tailor our protocols to your project needs, including target-specific protease digestion, labeling strategies, and choice of mass spectrometry method. This ensures optimal detection for your specific protein and research goals. -
Q: How experienced is your team?
A: Our lab is recognized globally for expertise in phosphorylation site mapping, with a track record of publications and collaborations. You benefit from a team that combines technical excellence with practical insights for your project.
Resources
Related Services
Related Products
References:
- Johnson JR, Santos SD, Johnson T, et al. Prediction of functionally important phospho-regulatory events in xenopus laevis oocytes. Radivojac P, ed. PLoS Comput Biol. 2015;11(8):e1004362. doi:10.1371/journal.pcbi.1004362
- Lanpher KB, Popendorf KJ.HPLC and 32P ‐radiolabeling method for quantification of microbial adenylate concentrations and turnover rates in seawater. Limnology & Ocean Methods. 2022;20(8):482-499. doi:10.1002/lom3.10499
- Plattner F, Bibb JA. Serine and threonine phosphorylation. In: Basic Neurochemistry. Elsevier; 2012:467-492. doi:10.1016/B978-0-12-374947-5.00025-0
- Zhang F, Qi L, Feng Q, et al. HIPK2 phosphorylates HDAC3 for NF-κB acetylation to ameliorate colitis-associated colorectal carcinoma and sepsis. Proc Natl Acad Sci USA. 2021;118(28):e2021798118. doi:10.1073/pnas.2021798118
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

