Chemokines
🧪 CXCL10-08H
Source: E.coli
Species: Human
Tag: Non
Conjugation:
Protein Length: 22-98 aa
$311.50
$623
/ 50 µg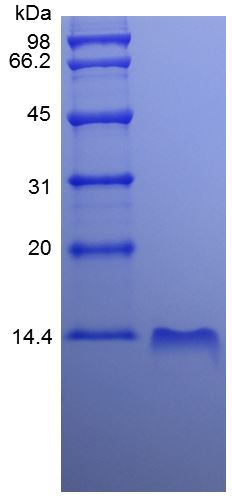
🧪 CCL1-19H
Source: Human Cells
Species: Human
Tag: Fc
Conjugation:
Protein Length: 24-96 aa

🧪 CXCL14-25H
Source: E.coli
Species: Human
Tag: His
Conjugation:
Protein Length:
🧪 CCL7-36H
Source: E.coli
Species: Human
Tag: Non
Conjugation:
Protein Length:

🧪 CCL7-46M
Source: E.coli
Species: Mouse
Tag: Non
Conjugation:
Protein Length:

🧪 CCL25-70H
Source: E.coli
Species: Human
Tag: Non
Conjugation:
Protein Length:
🧪 CCL5-85H
Source: Human Cells
Species: Human
Tag: His
Conjugation:
Protein Length: 1-91 a.a.
🧪 CX3CL1-86H
Source: HEK293
Species: Human
Tag: His
Conjugation:
Protein Length: Met1-Arg339
🧪 CCL11-138E
Source: Yeast
Species: Equine
Tag: Non
Conjugation:
Protein Length:
🧪 Cxcl16-153M
Source: HEK293
Species: Mouse
Tag: His
Conjugation:
Protein Length: Met1-Trp201

🧪 CXCL11-65S
Source: Yeast
Species: Pig
Tag: Non
Conjugation:
Protein Length:

🧪 CCL15-197H
Source: E.coli
Species: Human
Tag: His
Conjugation:
Protein Length: Gln22-Ile113
🧪 CCL17-192H
Source: Insect Cells
Species: Human
Tag: Non
Conjugation:
Protein Length: 1-94 aa

🧪 CCL17-193H
Source: Insect Cells
Species: Human
Tag: Non
Conjugation:
Protein Length: 1-94 a.a.
🧪 Ccl6-3993M
Source: HEK293
Species: Mouse
Tag: His
Conjugation:
Protein Length: Gly22-Ala116

Chemokines—Product Overview
Chemokines are a family of small signaling proteins that play a critical role in immune cell trafficking, inflammation and immune surveillance. By binding to specific G-protein-coupled receptors, chemokines regulate the migration and positioning of leukocytes and orchestrate immune responses in both physiological and pathological conditions. Due to their involvement in numerous diseases, including cancer, autoimmune and infectious diseases, chemokines are being extensively studied as potential therapeutic targets. Creative BioMart offers a comprehensive range of recombinant chemokines and related products to support research in immunology, inflammation and drug development.
Jump to Section
Product Families
-
CC Chemokines
-
CXC Chemokines
-
C Chemokines
-
CX3C Chemokines
-
Chemokine Receptors
Background
Chemokines are a family of small cytokines that regulate the migration and positioning of immune cells by binding to chemokine receptors, which are G protein-coupled receptors on target cells. They play a critical role in directing leukocytes to sites of infection, inflammation, and tissue injury. Chemokines also contribute to homeostatic immune surveillance by directing immune cells to lymphoid organs. Their precise regulation is essential for immune balance, and dysregulation has been implicated in several diseases, including chronic inflammation, autoimmune disorders and cancer.
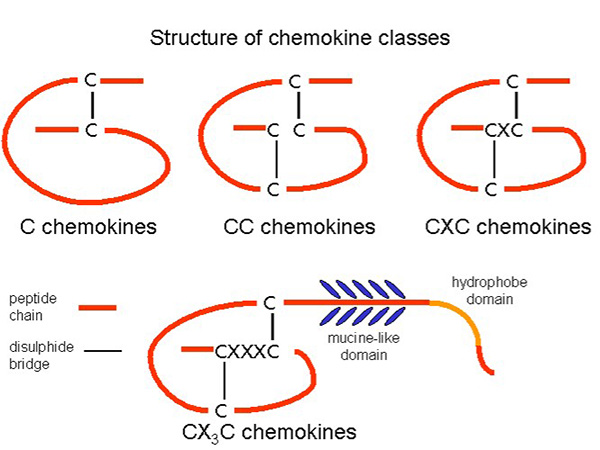
Chemokines are classified into four major subfamilies based on the arrangement of conserved cysteine residues: C, CC, CXC, and CX3C chemokines. CC chemokines primarily attract monocytes and T cells, whereas CXC chemokines often direct neutrophils. The CX3C subgroup, with its unique membrane-bound form, mediates both adhesion and migration. This classification helps define their specific roles in the immune response, with each subset influencing distinct leukocyte populations and inflammatory processes.
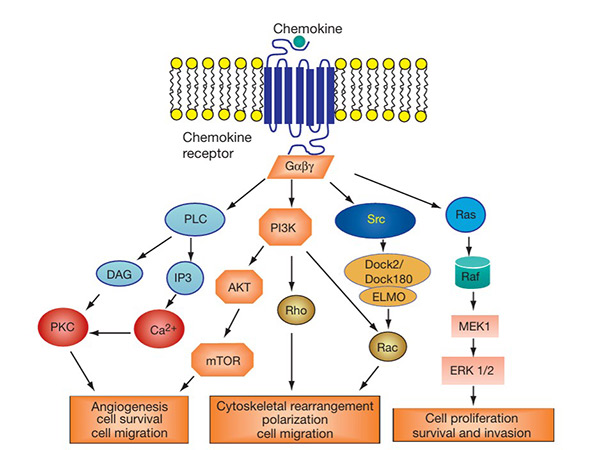
The chemokine signaling pathway is activated when chemokines bind to their corresponding GPCRs on target cells. This interaction triggers intracellular signaling cascades involving G proteins, leading to calcium mobilization, actin cytoskeleton rearrangement, and activation of kinases such as MAPK and PI3K. These signals regulate the migration, adhesion and survival of immune cells. Dysregulation of this signaling pathway is implicated in inflammatory diseases, autoimmune disorders and cancer. Targeting chemokine receptors in this pathway is a promising strategy for therapeutic intervention in various immune-related diseases.
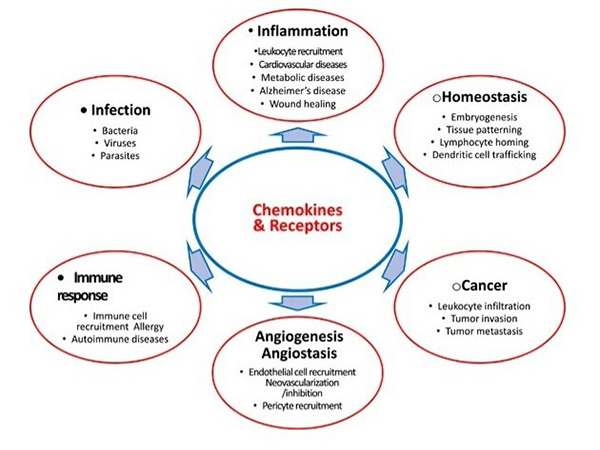
Chemokines contribute to various diseases by modulating immune responses, tumor progression and inflammatory disorders. In cancer, chemokines influence tumor growth, angiogenesis and metastasis. In autoimmune diseases, abnormal chemokine signaling leads to excessive immune cell infiltration and tissue damage. Because of their pivotal role, chemokines and their receptors are actively explored as therapeutic targets. Blocking or modulating chemokine activity holds great promise for treating diseases such as rheumatoid arthritis, multiple sclerosis and cancer, and several chemokine-based drugs are in clinical development.
Chemokines Receptors
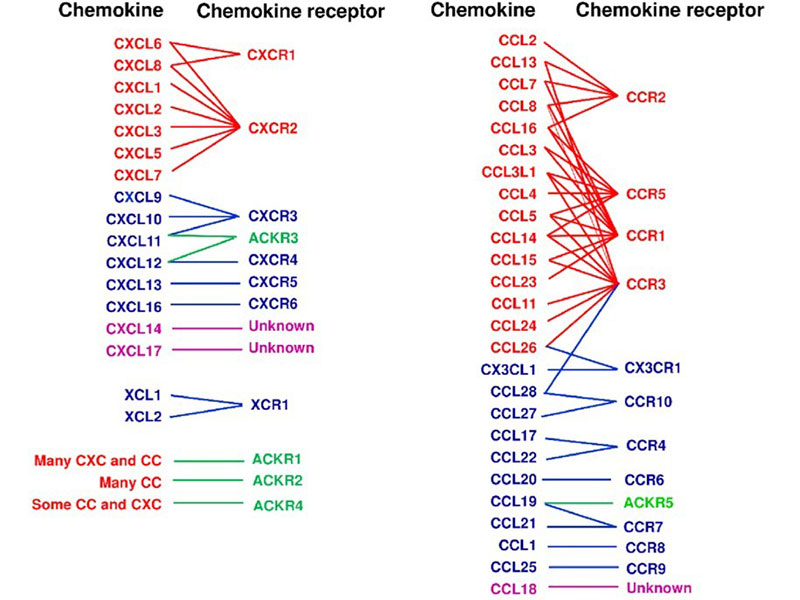
Chemokine receptors are a class of G protein-coupled receptors with seven transmembrane domains, primarily located on the surface of leukocytes. These receptors mediate the effects of chemokines by triggering intracellular signaling pathways that facilitate the migration of both inflammatory and non-inflammatory cells to specific tissues or compartments. To date, approximately 19 chemokine receptors have been identified, categorized into four main families based on their binding specificity: CXCR, which interacts with CXC chemokines; CCR, which binds CC chemokines; CX3CR1, which recognizes the unique CX3C chemokine (CX3CL1); and XCR1, which associates with the two XC chemokines (XCL1 and XCL2).
|
Human Chemokines |
Alternate Names |
Mouse Homologue |
Receptor |
|---|---|---|---|
|
CXC Chemokines |
|||
|
GROα, MGSA |
N51/KC, MIP-2 |
||
|
GROβ, M IP-2α |
Gro/KC |
||
|
GROγ, MIP-2β |
Gro/KC |
||
|
Platelet factor-4 |
None |
||
|
ENA-78 |
LIX |
||
|
GCP-2 |
LIX |
||
|
PBP |
None |
||
|
IL-8 |
None |
||
|
Mig |
Mig |
||
|
IP-10 |
CRG-2 |
||
|
I-TAC |
None |
||
|
SDF-Iα, SDF-Iβ |
SDF-Iα, SDF-Iβ |
||
|
BCA-1, BLC |
BLC |
||
|
BRAK, BMAC |
BRAK, BMAC |
Unknown |
|
|
None |
Lungkine |
Unknown |
|
|
CXCL16 |
CXCL16 |
||
|
VCC-1 |
CXCL17 |
Unknown |
|
|
CC Chemokines |
|||
|
I-309 |
TCA-3 |
||
|
MCP-1 |
JE |
||
|
MIP-1α, LD78α |
MIP-1α, LD78α |
||
|
MIP-1β |
MIP-1β |
||
|
RANTES |
RANTES |
||
|
None |
C10, MRP-1 |
Unknown |
|
|
MCP-3 |
MARC/FIC |
||
|
MCP-2 |
None |
||
|
None |
MRP-2, MIP-1γ |
Unknow |
|
|
Eotaxin-1 |
Eotaxin |
||
|
None |
MCP-5 |
||
|
MCP-4 |
None |
||
|
HCC-1 |
None |
||
|
HCC-2, leukotactin-1, MIP-5 |
None |
||
|
FICC-4, monotactin-1, LEC |
None |
Unknown |
|
|
TARC |
None |
||
|
DC-CK-1, PARC, MIP-4 |
None |
Unknown |
|
|
MIP-3β, ELC, exodus-3, ckβ11 |
None |
||
|
MIP-3α, LARC, exodus-1 |
None |
||
|
6-Ckine, SLC, exodus-2 |
6-Ckine, SLC |
||
|
MDC |
ABCD-1 |
||
|
MPIF-1, ckβ8 |
None |
||
|
MPIF-2, eotaxin-2, ckβ6 |
None |
||
|
TECK |
TECK |
Unknown |
|
|
Eotaxin-3, MIP-4α |
None |
||
|
Eskine |
ALP |
||
|
MEC |
CCL28/MEC |
||
|
C Chemokines |
|||
|
Lymophotactinα, SCM-1α |
lymophotactin |
||
|
XCL2 |
Lymophotactinβ, SCM-1β |
None |
|
|
CX3C Chemokines |
|||
|
Fractralkine, neurotactin |
Fractralkine, neurotactin |
||
Applications
Immunology and Inflammation Research
Recombinant chemokines are essential tools for studying immune cell migration, activation, and signaling in various inflammatory conditions. They help researchers investigate the role of chemokines in diseases such as rheumatoid arthritis, asthma, and inflammatory bowel disease. By using recombinant proteins, scientists can analyze chemokine-receptor interactions and develop targeted therapies aimed at modulating immune responses to treat chronic inflammatory and autoimmune disorders.
Cancer Research and Therapy Development
Chemokines play an important role in tumor progression, angiogenesis and metastasis. Recombinant chemokines allow researchers to study tumor microenvironment interactions, immune cell infiltration, and cancer cell migration. They are also used in drug screening to identify inhibitors of chemokine signaling that may prevent metastasis or enhance anti-tumor immunity. Targeting chemokine pathways is an emerging strategy for the development of novel immunotherapies, including checkpoint inhibitors and CAR-T cell therapy.
Infectious Disease Studies
Chemokines are critical for coordinating immune responses against viral, bacterial and parasitic infections. Recombinant chemokines allow researchers to study host-pathogen interactions, immune cell recruitment, and cytokine storms in diseases such as HIV, COVID-19, and tuberculosis. In addition, chemokine receptor antagonists are being explored as antiviral therapies, particularly in blocking viral entry mechanisms, as seen with CCR5 inhibitors in HIV treatment.
Drug Discovery and Development
Recombinant chemokines are widely used in pharmaceutical research to screen small molecules, antibodies and biologics that target chemokine signaling. They help to assess the efficacy of drugs in modulating immune responses, reducing inflammation, or inhibiting cancer progression. High-throughput screening platforms use recombinant chemokines to identify novel compounds that can serve as potential therapeutics for immune disorders, cancer, and infectious diseases, accelerating the development of targeted and personalized medicine.
Product Features
-
High Purity and Bioactivity: Our recombinant chemokines are produced using advanced expression systems to ensure high purity, proper folding, and optimal biological activity.
-
Wide Selection of Chemokines and Receptors: We offer a comprehensive range of recombinant chemokines and their receptors, covering the major CC, CXC, CX3C, and C families for diverse research applications.
-
Validated for Multiple Applications: Suitable for immunology, cancer research, drug discovery, and infectious disease studies, our chemokines are tested in cell-based assays, migration studies, and receptor binding experiments.
-
Customizable Solutions: Bulk production, custom formulations, and endotoxin-free options are available to meet specific research and biopharmaceutical development needs.
-
Stringent Quality Control Standards: Each batch undergoes rigorous testing including SDS-PAGE, HPLC, endotoxin assessment, and functional validation to ensure consistency and reliability.
-
Stringent Quality Control Standards: Each batch undergoes rigorous testing including SDS-PAGE, HPLC, endotoxin assessment, and functional validation to ensure consistency and reliability.
Case Study
Case 1: Chemokines in Tuberculous Lymphadenitis Patient
Novita et al., 2022. Characterization of chemokine and cytokine expression pattern in tuberculous lymphadenitis patient.
This study investigates the expression patterns of CCR-2, CCL-5, IL-6, IL-10, STAT-3, and SOCS-3 in tuberculous lymphadenitis. Using immunohistochemistry on 27 lymph node biopsy samples, researchers found that CCR-2, CCL-5, and IL-6 were highly expressed in lymphocytes and macrophages, while IL-10, STAT-3, and SOCS-3 had low expression. A strong correlation was observed between CCR-2 with IL-6 and IL-10. The findings suggest that chronic tuberculous lymphadenitis is characterized by high CCR-2, CCL-5, and IL-6 expression, with low STAT-3 and SOCS-3 levels, providing insights into the immune response and potential therapeutic targets in TB infection.
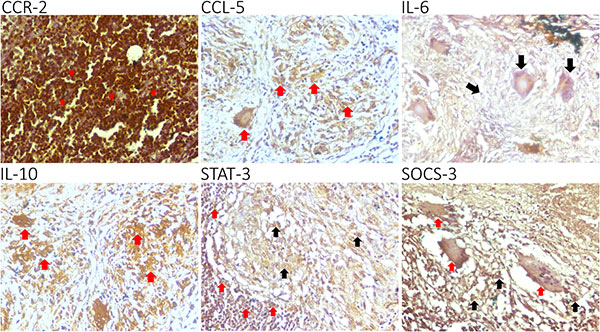
Figure 1. CCR-2, CCL-5, IL-6, IL-10, STAT-3, and SOCS-3 are expressed in the granuloma of a tuberculous lymphadenitis patient.
Case 2: CCL22-CCR4 Thermogenic Axis
Yuan et al., 2024. Macrophage-derived chemokine CCL22 establishes local LN-mediated adaptive thermogenesis and energy expenditure.
Lymph nodes (LNs) play a key role in promoting adipose beiging, particularly in inguinal white adipose tissue (iWAT). Removing local LNs impairs cold-induced beiging, which can be restored by injecting M2 macrophages or CCL22. CCL22 enhances iWAT beiging via its receptor CCR4, activating downstream signaling in eosinophils. Blocking CCL22 prevents beiging, while increasing its levels reduces body weight and fat mass. CCR4 inhibition with AZD2098 blocks CCL22-induced beiging, confirming the CCL22-CCR4 axis as a key regulator. These findings highlight CCL22 as a potential therapeutic target for obesity by promoting adaptive thermogenesis.
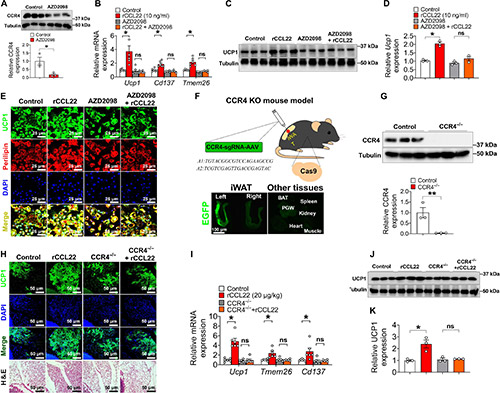
Figure 2. CCR4 is required for CCL22-induced iWAT beiging upon cold exposure.
FAQs
-
Q: What are chemokines, and why are they important in research?
A: Chemokines are a family of small cytokines that regulate the migration and signaling of immune cells. They play key roles in inflammation, immune surveillance, and disease pathology, making them critical for research in immunology, oncology, and infectious diseases. -
Q: What types of chemokine products do you offer?
A: We provide a wide range of recombinant chemokines, chemokine receptors, and assay kits designed for various research applications, including cell migration studies, signaling pathway analysis, and drug discovery.
-
Q: Are your chemokine products recombinant, and how are they produced?
A: Yes, our chemokines are recombinant proteins expressed in optimized host systems such as E. coli, mammalian, or insect cells to ensure high purity, proper folding, and bioactivity.
-
Q: How should chemokine proteins be stored?
A: Our recombinant chemokines should be stored at -20°C or -80°C for long-term stability. For short-term use, we recommend storage at 4°C to maintain activity. Please refer to the product data sheet for specific storage conditions. -
Q: Do your chemokines have endotoxin specifications?
A: Yes, we offer endotoxin-tested chemokine proteins with endotoxin levels below industry standards, ensuring suitability for in vitro and in vivo research applications. -
Q: Can I request bulk orders or custom formulations?
A: Absolutely! We offer bulk supply options and custom formulations to meet specific research needs. Contact our team to discuss your requirements.

