Protein Pathway Profiling
Background
Cellular function is governed by complex networks of signaling pathways that control everything from growth and differentiation to immune responses and cell death. These pathways are orchestrated through dynamic protein interactions, phosphorylation events, and regulatory feedback loops. When disrupted, such pathways often contribute to disease progression, including cancer, autoimmune disorders, and neurodegenerative diseases.
Protein pathway profiling is a powerful tool that enables researchers to investigate these signaling cascades in detail. By simultaneously monitoring multiple proteins and their phosphorylation states, scientists can gain a systems-level understanding of cellular responses to drugs, environmental changes, or genetic alterations.
At Creative BioMart, our Protein Pathway Profiling Service enables high-throughput, quantitative analysis of key signaling events using our library of highly specific antibodies. These tools allow precise detection of dysregulation and phosphorylation status across critical cell signaling pathways—insights essential for drug discovery and therapeutic development. Whether you're investigating pathway activation, drug mechanisms, or identifying biomarkers, our platform delivers the accuracy and biological context needed to drive impactful research.
What We Offer?
Service Procedure

Applications of our Services
Drug Discovery & Development
Protein pathway profiling supports early-stage drug discovery by enabling the evaluation of mechanism of action, dose response, and time-course effects in cell-based assays. It also aids in the assessment of combination therapies and detection of off-target effects, helping streamline the development of safer, more effective therapeutics.
Biological Research & Disease Mechanism Studies
Gain insight into dysregulated signaling pathways in disease models using biological samples or fixed tissues. This application is critical for understanding disease progression, identifying key therapeutic targets, and validating in vitro or animal models for translational research.
Resistance Mechanisms & Biomarker Identification
Identify pathways involved in drug resistance and uncover predictive or prognostic biomarkers that can guide patient selection, therapeutic decisions, or disease staging.
Biologics & Biosimilar Characterization
Compare the functional signaling fingerprints of biologics and biosimilars to ensure consistency in activity, helping meet regulatory standards and support product development.
Why Choose Us?
- Comprehensive Pathway Coverage: We analyze a wide range of signaling cascades—including MAPK, PI3K/Akt, JAK/STAT, and NF-κB—to reveal intricate pathway dynamics.
- Customizable Assay Panels: Our pathway profiling panels are fully customizable for specific targets, cell types, disease models, or treatment conditions.
- Quantitative, High-Throughput Analysis: We deliver precise, multiplexed data using advanced platforms like Luminex, MSD, or Western blotting for high-throughput applications.
- Mechanistic Insight Support: Our service links pathway activity with cellular responses to help interpret upstream signals and downstream functional effects.
- Biologically Relevant Models: We work with primary cells, disease-relevant lines, or 3D cultures to ensure data reflect in vivo-like conditions.
- Expert Data Interpretation: Every project includes detailed data reporting and scientific consultation to contextualize findings within your therapeutic or research objectives.
Case Study
* NOTE: We prioritize confidentiality to safeguard our clients’ technology and intellectual property. As an alternative, we present selected published research articles as representative case studies. For details on the assay services and products used in these studies, please refer to the relevant sections of the cited literature.
Case 1: Molecular profiling of PI3K-AKT-MTOR variants in focal cortical malformations
Pirozzi et al., 2022. doi:10.1093/brain/awab376
This study investigated PI3K-AKT-MTOR pathway mutations in children with focal malformations of cortical development, including focal cortical dysplasia and hemimegalencephaly—conditions linked to epilepsy, autism, and intellectual disability. Using ultra-sensitive droplet digital PCR, researchers screened 159 samples from 58 patients for six known hotspot mutations. Pathogenic variants were found in 29.31% of cases, with MTOR mutations prevalent in cortical dysplasia and PIK3CA mutations more common in hemimegalencephaly. Although mosaicism levels varied, they did not correlate with malformation severity. These findings support ddPCR as a valuable diagnostic tool for PI3K-AKT-MTOR-related brain disorders.
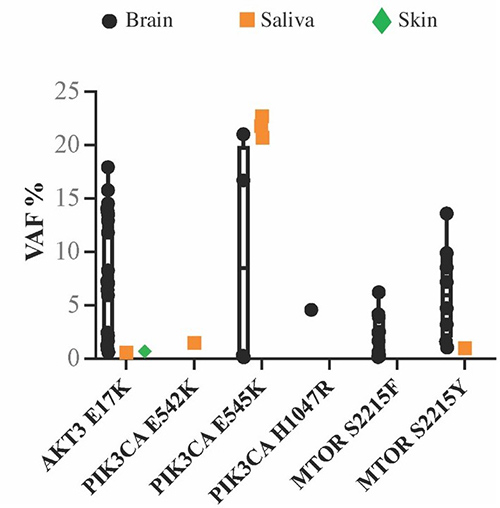
Figure 1. Positive samples stratified by the PI3K-AKT-MTOR hotspot mutation to visualize the range of VAF for each mutation across individuals. Notably, the PIK3CA, p.E545K, variant had the widest VAF% range and was the only hotspot to be present at higher VAF% in saliva rather than brain samples. (Pirozzi et al., 2022)
Case 2: Ganoderma sterols suppress inflammation via p38 and NF-κB pathways
Xu et al., 2021. doi:10.1016/j.fct.2021.112073
This study demonstrates that sterols from Ganoderma lucidum (GLS), isolated through an efficient single-step method, significantly contribute to the fungus's anti-inflammatory effects. GLS reduced LPS-induced inflammation in macrophages by decreasing pro-inflammatory mediators (NO, TNF-α, IL-1β, IL-6) at both protein and mRNA levels. Mechanistically, GLS inhibited the MAPK pathway by blocking p38 phosphorylation and suppressed NF-κB activation by preventing IκB-α degradation and p65 phosphorylation. Molecular docking confirmed direct sterol binding to p38 and p65, suggesting GLS as promising, natural anti-inflammatory agents.
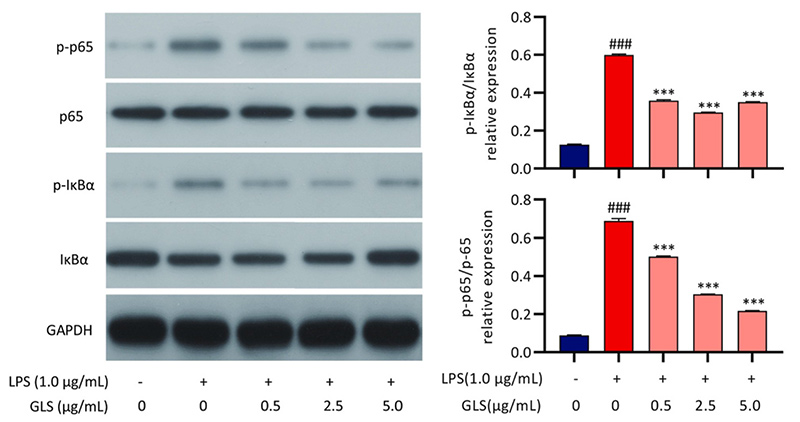
Figure 2. Effects of GLS on the LPS-stimulated activation of NF-kB signaling pathways in RAW264.7 cells. Cells were pretreated with GLS (0.5, 2.5 and μg/mL) for 2 h and then treated with 0.1 μg/mL LPS for 1 h. The levels of p-IκBα, IκBα, p-p65 and p65 were detected by immunoblotting with total cell lysates. (Xu et al., 2021)
Customer Testimonials
-
"We used Creative BioMart’s PI3K/Akt pathway profiling to assess compound effects in tumor cell lines. The data were clear, reproducible, and instrumental in advancing our lead candidate."
— Senior Scientist | Biotech R&D Division
-
"Creative BioMart’s customized JAK/STAT pathway analysis in primary immune cells helped us pinpoint the downstream effects of cytokine modulation. Outstanding technical support throughout."
— Principal Investigator | Academic Medical Center
-
"The pathway profiling service from Creative BioMart provided us with a comprehensive overview of MAPK signaling under drug treatment. The turnaround time was fast, and the interpretation was accurate."
— Director of Preclinical Pharmacology | Mid-sized Pharma
-
"We relied on phospho-protein analysis from Creative BioMart to evaluate NF-κB activation in response to TLR ligands. The team also helped us optimize our assay conditions."
— Lead Researcher | Immuno-Oncology Startup
-
"Creative BioMart provided a thorough multiplex readout for multiple signaling cascades in 3D tumor models. These insights significantly influenced our biomarker strategy."
— Project Manager | Translational Research Group
FAQs
-
Q: What types of signaling pathways can you profile?
A: We offer comprehensive profiling of major signaling cascades, including MAPK/ERK, PI3K/Akt, JAK/STAT, NF-κB, TGF-β, and more. Panels can be customized for your targets. -
Q: What detection platforms do you use for pathway profiling?
A: Depending on your needs, we use phospho-specific Western blotting, multiplex bead-based assays (e.g., Luminex), MSD platforms, and high-content imaging. -
Q: How customizable are your assay panels?
A: Highly customizable. You can select specific phospho-proteins, total proteins, or pathway-specific readouts. We tailor the assay to your research question. -
Q: Do you help interpret the data?
A: Absolutely. Our scientific team provides full analysis, including pathway maps, quantification, and biological interpretation aligned with your experimental goals. -
Q: What’s your typical turnaround time?
A: Most projects are completed within 2–3 weeks, depending on complexity. Expedited services are available upon request.
Resources
Related Services
- Protein Labeling
- Protein Characterization
- Protein Analytical Service
- Protein Interaction Service
- Protein Microarray Service
- Custom Protein Service
- Enzyme Activity Assay
Related Products
References:
- Pirozzi F, Berkseth M, Shear R, et al. Profiling PI3K-AKT-MTOR variants in focal brain malformations reveals new insights for diagnostic care. Brain. 2022;145(3):925-938. doi:10.1093/brain/awab376
- Xu J, Xiao C, Xu H, et al. Anti-inflammatory effects of Ganoderma lucidum sterols via attenuation of the p38 MAPK and NF-κB pathways in LPS-induced RAW 264.7 macrophages. Food and Chemical Toxicology. 2021;150:112073. doi:10.1016/j.fct.2021.112073
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

