Epigenetics
Creative BioMart Epigenetics Product List
Immunology Background
Background
Epigenetics refers to the study of heritable changes in gene expression that do not involve alterations to the underlying DNA sequence. In classical genetics, traits pass from generation to generation in DNA, the strands of genetic material that encode your genes. Scientists thought alterations to the DNA itself was the only way changes could pass on to subsequent generations.
Some phenomena of heredity cannot be explained by the DNA mode of classical genetics: 1. During cell differentiation, daughter cells with different functions are created in the course of cell division, although the genetic material is the same in all cells. Determining the functional identity of a cell is a topic of epigenetics. 2. There are properties that are only "inherited" from the father (paternal), just as there are properties that only originate from the mother (maternal) and are not related to the base sequence. 3. When functionally defined cells (terminally differentiated cells) are converted back into undifferentiated cells, which can develop into different cells again and which are used in the cloning of individuals (e.g. Dolly), epigenetic fixations must be removed so that a cell does not remain fixed to a single function but can again acquire and inherit all or many functions.
Epigenetics has revolutionized our understanding of genetics by revealing how genes can be turned on or off and how this regulation affects cellular function and organismal development. The term "epigenetics" originates from the Greek word "epi," meaning "above" or "on top of" genetics, signifying that these changes are layered over the genetic code. Unlike genetic mutations, which alter the DNA sequence itself, epigenetic modifications affect how cells read genes, providing an additional layer of control over genetic information.
Mechanisms of Epigenetic Regulation
The primary mechanisms of epigenetic regulation include DNA methylation, histone modification, and non-coding RNA-associated gene silencing. Each of these mechanisms can significantly impact gene expression and cellular function.
DNA Methylation
This involves the addition of a methyl group to the DNA molecule, typically at cytosine bases that are followed by guanine (CpG sites). DNA methylation generally acts to repress gene transcription. Thus, environmental factors can impact the amount of protein a cell makes. Less protein might be made if an environmental factor causes an increase in DNA methylation, and more protein might be made if a factor causes an increase in demethylation. In mammalian cells, DNA methylation patterns are crucial for normal development and are associated with processes such as X-chromosome inactivation and imprinting.
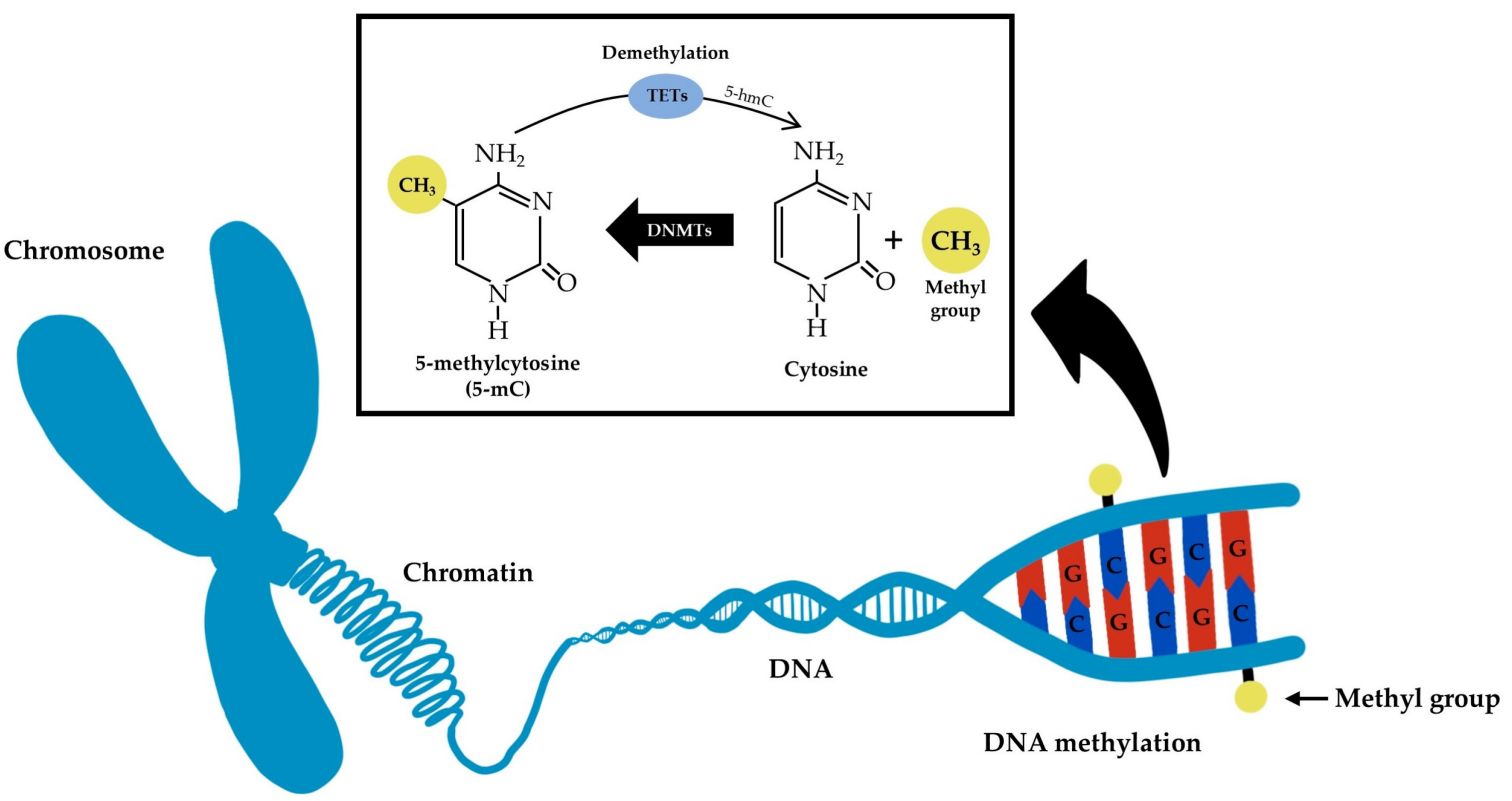 Fig. 1: Simplified schematic representation of DNA methylation and demethylation molecular mechanisms.
Fig. 1: Simplified schematic representation of DNA methylation and demethylation molecular mechanisms.Histone Modification
Histones are proteins around which DNA winds, forming a complex called chromatin. The modification of histones, such as acetylation, methylation, phosphorylation, and ubiquitination, can influence chromatin structure and gene expression. For instance, acetylation of histone tails by histone acetyltransferases (HATs) is typically associated with transcriptional activation, while deacetylation by histone deacetylases (HDACs) leads to transcriptional repression.
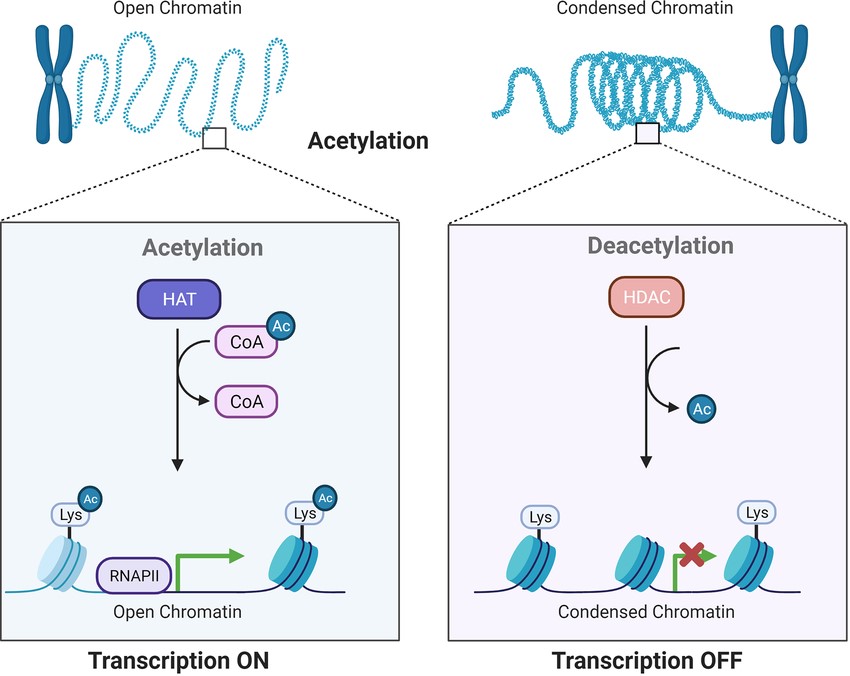 Fig. 2: Acetylation by HATs and deacetylation by HDACs influence gene transcriptional activity. HATs and HDACs add or remove acetyl groups at the N-terminus lysine, which leads to an open or condensed state of the chromatin.
Fig. 2: Acetylation by HATs and deacetylation by HDACs influence gene transcriptional activity. HATs and HDACs add or remove acetyl groups at the N-terminus lysine, which leads to an open or condensed state of the chromatin.Non-Coding RNAs
These are RNA molecules that are not translated into proteins but can regulate gene expression at the transcriptional and post-transcriptional levels. Examples include microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). MicroRNAs function primarily by binding to complementary sequences on target messenger RNAs (mRNAs), leading to mRNA degradation or inhibition of translation. This post-transcriptional regulation is a key component of the cellular machinery that ensures proper gene expression patterns. lncRNAs are transcribed by RNA polymerase II from various genomic regions, including intergenic, intronic, and enhancer regions. Although the proportion of functional lncRNAs is still not clear, many lncRNAs play important regulatory roles in diverse biological processes. Besides their participation to normal physiology, lncRNA expression and function have been already associated to many diseases, including cancer. By interacting with epigenetic regulators and by controlling chromatin topology, their misregulation may result in an aberrant regulation of gene expression that may contribute to tumorigenesis.
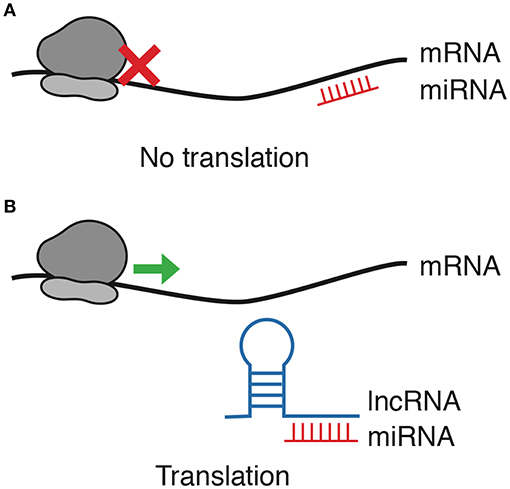 Fig. 3: lncRNA/miRNA/mRNA axis regulation. (A) miRNAs block translation by binding mRNA. (B) As lncRNAs function as decoys for miRNAs, mRNA translation is allowed.
Fig. 3: lncRNA/miRNA/mRNA axis regulation. (A) miRNAs block translation by binding mRNA. (B) As lncRNAs function as decoys for miRNAs, mRNA translation is allowed.Influences on Epigenetic Modifications
Epigenetic modifications are influenced by a variety of internal and external factors, including developmental cues, environmental exposures, lifestyle factors, and stochastic events.
Developmental Influences
During development, epigenetic mechanisms ensure that genes are expressed in a time- and tissue-specific manner. For instance, during embryogenesis, certain genes are activated or silenced to direct the differentiation of stem cells into various cell types. A prime example of this is the differentiation of muscle and nerve cells. Despite having the same genetic material, these cells develop into functionally and structurally distinct cell types due to epigenetic modifications.
Environmental Exposures
Environmental factors such as diet, toxins, stress, and physical activity can lead to changes in the epigenome. For example, exposure to pollutants like heavy metals and endocrine disruptors has been linked to aberrant DNA methylation and increased risk of diseases.
Diet and nutrition have significant effects on the epigenome. Nutrients such as folate, vitamins B12, B6, and choline are sources of methyl groups required for DNA methylation. A diet deficient in these nutrients can lead to hypomethylation and dysregulation of gene expression. Particularly nutrition during pregnancy have been paid attention to. Studies have shown that during the Dutch Hunger Winter famine of 1944-1945, people whose mothers were pregnant with them during the famine were more likely to develop certain diseases, including heart disease, schizophrenia, and type 2 diabetes. About 60 years after the famine, researchers looked at DNA methylation levels in people whose mothers were pregnant with them during the famine. These people had increased DNA methylation at some genes and decreased DNA methylation at others, compared with their siblings who were not exposed to famine before birth. These differences in DNA methylation may help explain why these people had an increased likelihood of certain diseases later in life.
Epigenetics and Diseases
Aberrant epigenetic modifications are implicated in a wide range of diseases, including cancer, neurological disorders, cardiovascular diseases, and metabolic disorders.
Cancer
Cancer is often associated with global changes in DNA methylation patterns, including hypermethylation of tumor suppressor genes and hypomethylation of oncogenes. These alterations can disrupt normal cellular regulation and contribute to tumorigenesis.
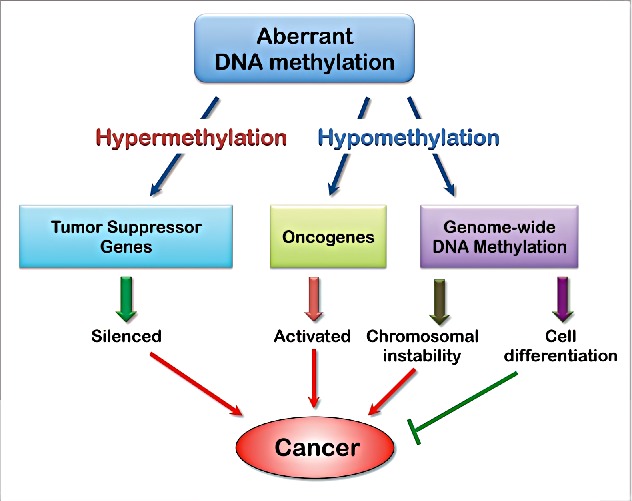 Fig. 4: Aberrant DNA methylation contributes to tumorigenesis (A). Hypermethylation of the promoter of tumor suppressor genes (TSGs) or gene family leads to its silencing and contributes to tumorigenesis. (B) Hypomethylation of oncogene promoters leads to their activation and contributes to tumorigenesis. (C) Genome-wide DNA hypomethylation may lead to chromosomal instability or cell differentiation, which contributes to tumorigenesis or inhibits tumorigenesis, respectively.
Fig. 4: Aberrant DNA methylation contributes to tumorigenesis (A). Hypermethylation of the promoter of tumor suppressor genes (TSGs) or gene family leads to its silencing and contributes to tumorigenesis. (B) Hypomethylation of oncogene promoters leads to their activation and contributes to tumorigenesis. (C) Genome-wide DNA hypomethylation may lead to chromosomal instability or cell differentiation, which contributes to tumorigenesis or inhibits tumorigenesis, respectively.A key aspect of these changes is transcription silencing, where the hypermethylation of promoter regions in tumor suppressor genes leads to their inactivity, preventing them from controlling cell growth and division. For example, the tumor suppressor gene p16INK4a, which plays a critical role in regulating the cell cycle, is often found hypermethylated and silenced in various cancers, including melanoma, lung cancer, cervical cancer, and pancreatic cancer. This silencing removes an essential checkpoint in cell cycle control, allowing unchecked cellular proliferation.
Conversely, hypomethylation of oncogenes, which normally would be tightly regulated, can lead to their overexpression and contribute to cancer progression. For instance, the R-Ras oncogene, which is involved in cell growth and differentiation, can become hypomethylated and thus overexpressed in certain types of cancers, such as gastric and colorectal cancers. This overexpression drives the malignant behavior of cancer cells, enhancing their proliferative and invasive capabilities.
Neurological Disorders
Epigenetic mechanisms are crucial for brain development and function. Dysregulation of these processes is linked to neurological disorders such as Alzheimer's disease, schizophrenia, and autism spectrum disorders. For example, several epigenetic mechanisms contribute to Alzheimer's dysregulation. In the healthy brain, there are a number of epigenetic modifications thar occur on histone tails, such as acetylation, methylation, and phosphorylation. In AD brains, however, gains or losses of histone modifications can be observed.
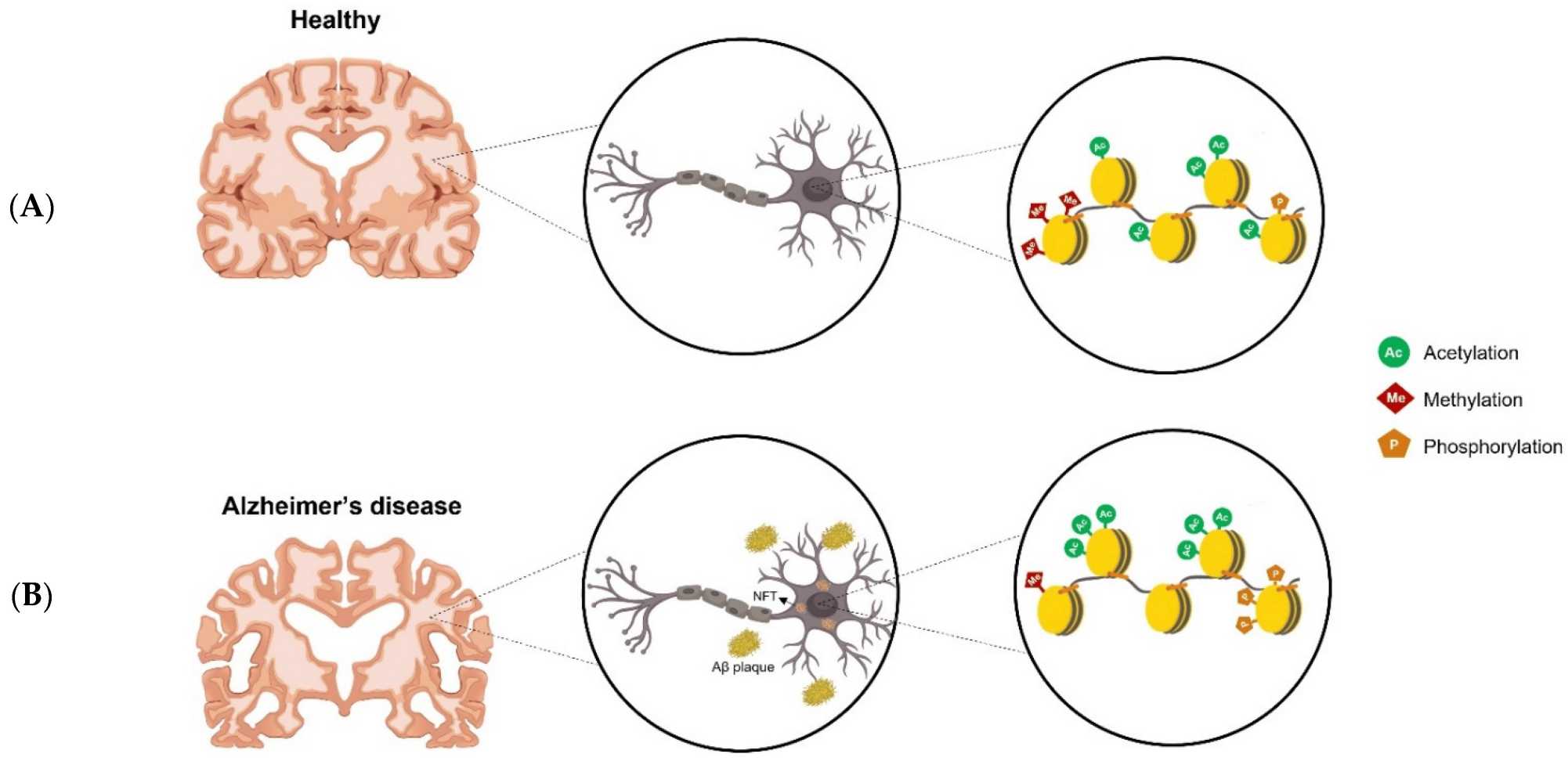 Fig. 5: Dysregulation of epigenetic modifications in AD. (A) A healthy brain with normal epigenetic modifications. (B) Increased or decreased histone modifications in the AD's brain.
Fig. 5: Dysregulation of epigenetic modifications in AD. (A) A healthy brain with normal epigenetic modifications. (B) Increased or decreased histone modifications in the AD's brain.Cardiovascular Diseases
Epigenetic changes can influence the development of cardiovascular diseases by affecting genes involved in inflammation, lipid metabolism, and endothelial function. For example, DNA methylation of genes related to vascular function has been associated with atherosclerosis. Hypermethylation of the promoter region of the eNOS (endothelial nitric oxide synthase) gene, which is crucial for maintaining vascular tone and health, has been observed in atherosclerotic plaques. This hypermethylation reduces eNOS expression, leading to decreased nitric oxide production, endothelial dysfunction, and increased susceptibility to plaque formation and vascular inflammation.
Metabolic Disorders
Epigenetic modifications play a role in the regulation of metabolic pathways and can contribute to disorders such as obesity and diabetes. Changes in DNA methylation and histone modifications can affect the expression of genes involved in glucose and lipid metabolism, influencing disease susceptibility. For instance, altered DNA methylation patterns in the promoter region of the PPARγ (peroxisome proliferator-activated receptor gamma) gene, which is critical for adipogenesis and lipid metabolism, have been observed in obese individuals. Hypermethylation of PPARγ can reduce its expression, impairing lipid storage and leading to an increased risk of developing obesity and insulin resistance.Therapeutic Potential of Epigenetics
Understanding epigenetic mechanisms opens up new avenues for therapeutic interventions. Epigenetic therapies aim to reverse abnormal gene expression patterns by targeting enzymes involved in DNA methylation and histone modification.
DNA Methylation Inhibitors
Drugs such as 5-azacytidine and decitabine are DNA methylation inhibitors used in the treatment of certain types of cancer. These drugs incorporate into DNA and inhibit DNMTs, leading to the reactivation of silenced tumor suppressor genes.
Histone Deacetylase Inhibitors
Histone deacetylase inhibitors (HDAC inhibitors) are another class of epigenetic drugs that promote a more relaxed chromatin structure, enhancing the expression of genes involved in cell cycle regulation and apoptosis. HDAC inhibitors are being explored for the treatment of cancer and other diseases.
 Fig. 6: Epigenetics has therapeutic potential. Improving health through medication, exercise, and diet.
Fig. 6: Epigenetics has therapeutic potential. Improving health through medication, exercise, and diet.Case Study
Case 1: Chioccioli, M.; et al. A lung targeted miR-29 mimic as a therapy for pulmonary fibrosis. EBioMedicine. 2022 Nov;85:104304.
MicroRNAs are non-coding RNAs that negatively regulate gene networks. The miR-29 family targets DNMT3A and DNMT3B, enzymes involved in DNA methylation. By downregulating these DNMTs, miR-29 can lead to global hypomethylation and reactivation of silenced genes, including tumor suppressor genes in cancer cells. Previous studies have shown that systemically delivered miR-29 mimic MRG-201 reduced fibrosis in animal models, supporting the consideration of miR-29-based therapies for idiopathic pulmonary fibrosis (IPF). Researchers generated MRG-229, a next-generation miR-29 mimic based on MRG-201 conjugated with peptide. The results demonstrated that the MRG-229 shows robust anti-fibrotic activity in vivo.
To assess in vivo activity of MRG-229, a prophylactic paradigm was used in the bleomycin-induced pulmonary fibrosis mouse model, in which we administered compound at day 3 following bleomycin administration and collected tissue at day 14. At day 14, Chioccioli et al. found a comparable down-regulation of miR-29 direct targets as well as non-direct targets (i.e., CTGF) in MRG-201 and MRG-229 bleomycin-injured lungs. Prophylactic doses of MRG-201 (100mpk) and peptide-coupled MRG-229 mimicry (10mpk) were analyzed on day 14.
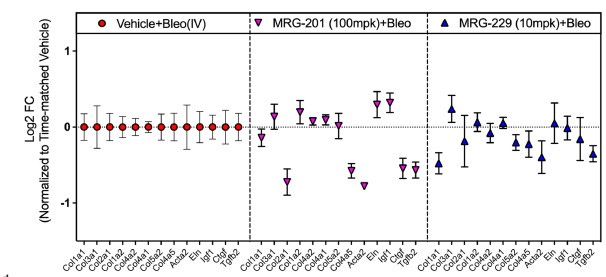 Fig. 7: qPCR analysis of downregulated gene expression levels of a panel of fibrosis-associated genes in lung harvested from bleomycin-induced mice treated with either MRG-201 or MRG-229.
Fig. 7: qPCR analysis of downregulated gene expression levels of a panel of fibrosis-associated genes in lung harvested from bleomycin-induced mice treated with either MRG-201 or MRG-229.Case 2: Oubaddou, Y.; et al. BRCA1 promoter hypermethylation in malignant breast tumors and in the histologically normal adjacent tissues to the tumors: exploring its potential as a biomarker and its clinical significance in a translational approach. Genes, vol. 14, no. 9, Sept. 2023, p. 1680.
The hypermethylation status of the promoter region of the breast cancer 1 (BRCA1), a well-known tumor suppressor gene, has been extensively investigated in the last two decades as a potential biomarker for breast cancer. BRCA1 is involved in DNA repair, and its inactivation through methylation-induced silencing can lead to genetic instability and an increased risk of tumor development. In this study, researchers used methylation-specific PCR (MSP) to analyze BRCA1 promoter hypermethylation in malignant breast tumors (MBTs), normal adjacent tissues (NATs), and benign breast lesions (BBLs). The results showed that BRCA1 promoter hypermethylation was higher in MBTs and NATs compared to BBLs. The high percentage of BRCA1 hypermethylation in the histologically normal adjacent tissues to the tumors (NATs) suggests the involvement of this epigenetic silencing as a potential biomarker of the early genomic instability in NATs surrounding the tumors.
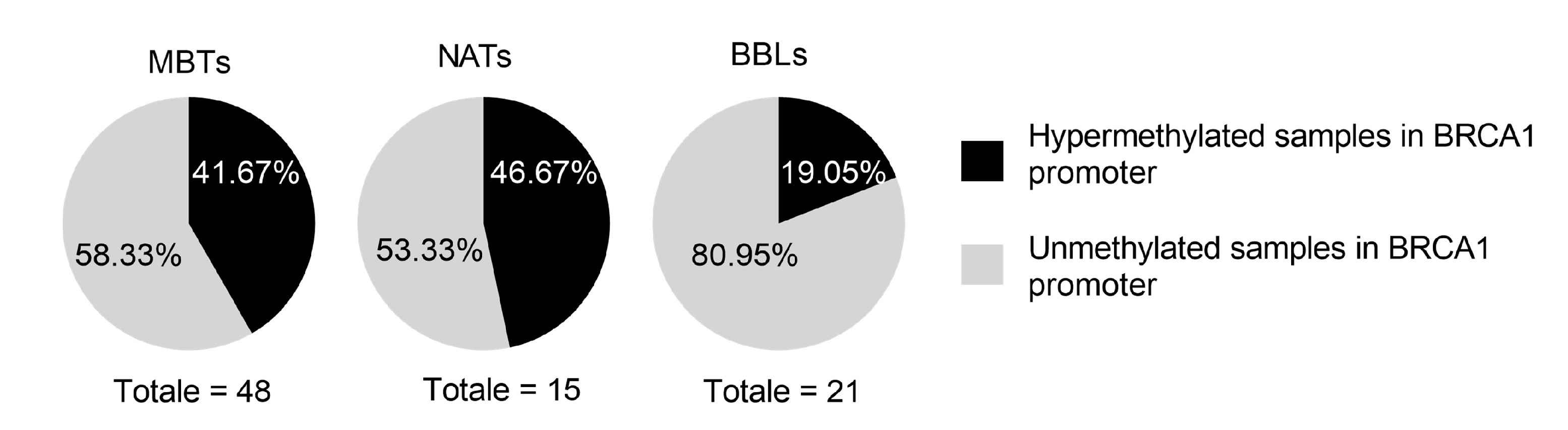 Fig. 8: Results of methylation-specific PCR (MSP) analysis of the BRCA1 promoter in 84 FFPE samples. Portions of BRCA1 promoter hypermethylation and unmethylation in different breast tissue samples: Malignant Breast Tumors (48MBTs), Normal Adjacent Tissues (15NATs), and Benign Breast Lesions (21BBLs).
Fig. 8: Results of methylation-specific PCR (MSP) analysis of the BRCA1 promoter in 84 FFPE samples. Portions of BRCA1 promoter hypermethylation and unmethylation in different breast tissue samples: Malignant Breast Tumors (48MBTs), Normal Adjacent Tissues (15NATs), and Benign Breast Lesions (21BBLs).References
- Chioccioli, Maurizio, et al. "A Lung Targeted miR-29 Mimic as a Therapy for Pulmonary Fibrosis." EBioMedicine, vol. 85, Nov. 2022, p. 104304. PubMed, https://doi.org/10.1016/j.ebiom.2022.104304.
- Hai, Rihan, et al. "Characterization of Histone Deacetylase Mechanisms in Cancer Development." Frontiers in Oncology, vol. 11, 2021, p. 700947. PubMed, https://doi.org/10.3389/fonc.2021.700947.
- López-Urrutia, Eduardo, et al. "Crosstalk Between Long Non-Coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer." Frontiers in Oncology, vol. 9, July 2019. Frontiers, https://doi.org/10.3389/fonc.2019.00669.
- Morlando, Mariangela, and Alessandro Fatica. "Alteration of Epigenetic Regulation by Long Noncoding RNAs in Cancer." International Journal of Molecular Sciences, vol. 19, no. 2, Feb. 2018, p. 570. PubMed Central, https://doi.org/10.3390/ijms19020570.
- Oubaddou, Yassire, et al. "BRCA1 Promoter Hypermethylation in Malignant Breast Tumors and in the Histologically Normal Adjacent Tissues to the Tumors: Exploring Its Potential as a Biomarker and Its Clinical Significance in a Translational Approach." Genes, vol. 14, no. 9, Sept. 2023, p. 1680. www.mdpi.com, https://doi.org/10.3390/genes14091680.
- Painter, Rc, et al. "Transgenerational Effects of Prenatal Exposure to the Dutch Famine on Neonatal Adiposity and Health in Later Life." BJOG: An International Journal of Obstetrics & Gynaecology, vol. 115, no. 10, Sept. 2008, pp. 1243–49. DOI.org (Crossref), https://doi.org/10.1111/j.1471-0528.2008.01822.x.
- Santana, Dalileia Aparecida, et al. "Histone Modifications in Alzheimer's Disease." Genes, vol. 14, no. 2, Feb. 2023, p. 347. www.mdpi.com, https://doi.org/10.3390/genes14020347.
- Valente, Ana, et al. "The Effect of Nanomaterials on DNA Methylation: A Review." Nanomaterials, vol. 13, no. 12, Jan. 2023, p. 1880. www.mdpi.com, https://doi.org/10.3390/nano13121880.
- Winblad, S. "Erythema Nodosum Associated with Infection with Yersinia Enterocolitica." Scandinavian Journal of Infectious Diseases, vol. 1, no. 1, May 1969, pp. 11–16. Europe PMC, https://doi.org/10.3109/inf.1969.1.issue-1.02.

