Drug Analysis Service
Background
Drug analysis is a critical component in the development and evaluation of pharmaceutical compounds, particularly during the pre-clinical and clinical phases of drug discovery. It involves the comprehensive examination of a drug's physical and chemical properties, as well as its behavior within biological systems. Accurate and reliable drug analysis is essential for ensuring the safety, efficacy, and quality of pharmaceutical products, from early-stage research to final market approval.
The process of drug development is highly complex and regulated, requiring rigorous testing and validation at multiple stages. Drug analysis plays a pivotal role in this process by providing detailed insights into the characteristics and behavior of drug candidates. This information is crucial for optimizing drug formulations, understanding pharmacokinetics (how the drug moves through the body), and assessing potential toxicological risks. Ultimately, drug analysis helps researchers and pharmaceutical companies make informed decisions, streamline the development process, and bring safe and effective medications to market.

Creative BioMart provides Drug Analysis Service for pre-clinical drugs and natural products on their physical and chemical properties. Our services include three main assays: pre-clinical pharmaceutical property analysis , pharmacokinetics analysis & bioanalysis , and natural product analysis .
Overview of Our Drug Analysis Service
Service Procedure

Service Details
|
Category |
 Pre-Clinical Pharmaceutical Property Analysis |
 Pharmacokinetics Analysis & Bioanalysis |
 Natural Product Analysis |
|
Description |
The presence of impurities in pharmaceutical compounds can significantly impact drug safety, stability, and efficacy. These impurities may arise during the production process or as a result of storage conditions such as humidity and temperature. Our comprehensive pre-clinical pharmaceutical property analysis service is designed to detect, identify, and quantify these impurities, thereby ensuring the quality and safety of drug candidates. |
A thorough understanding of a compound’s pharmacokinetic properties is essential for the successful development of therapeutics. We offer a complete suite of in vitro and in vivo pharmacokinetics and bioanalytical services to support drug discovery and development efforts. |
Natural products represent a rich source of bioactive compounds with therapeutic potential. However, their complex composition presents unique analytical challenges. Our specialized services in natural product analysis are designed to elucidate composition, ensure quality control, and support mechanism-of-action studies. |
|
Service Details |
|
In Vitro ADME (Absorption, Distribution, Metabolism, and Elimination):
In Vivo Pharmacokinetics and Bioanalysis:
|
|
|
Techniques and Instruments |
|
|
|
Advantages of Our Drug Analysis Service
- Full-Spectrum Services : From impurity profiling to pK and natural product analysis—everything under one roof.
- Cutting-Edge Instruments : High-sensitivity data from advanced LC-MS/MS, NMR, HRMS, and GC-MS platforms.
- Impurity & Stability Expertise : Accurate detection and structural ID of impurities and degradants, with ICH-compliant testing.
- In-Depth ADME/PK Studies : LogD, pKa, enzyme profiling, and in vivo rodent studies for complete pharmacokinetic insight.
- Specialized Natural Product Support : Metabolomics and multi-component quantification tailored for botanical and herbal compounds.
- Regulatory-Ready Methods : Custom method development and validation for compliant, submission-grade results.
Case Studies on Pharmacokinetics and Natural Product Analysis
* NOTE: We prioritize confidentiality to safeguard our clients’ technology and intellectual property. As an alternative, we present selected published research articles as representative case studies. For details on the assay services and products used in these studies, please refer to the relevant sections of the cited literature.
Case 1: Study of lipophilicity and ADME properties of 1,9-diazaphenothiazines with anticancer action
Morak-Mdt>ł odawska et al ., 2023. doi:10.3390/ijms24086970
This study focuses on the lipophilicity and bioavailability of 10-substituted 1,9-diazaphenothiazines, compounds with notable in vitro anticancer potential due to their activation of the mitochondrial apoptosis pathway. Researchers assessed lipophilicity both theoretically using computational tools and experimentally through reverse-phase thin-layer chromatography (RP-TLC). Additional evaluations included physicochemical, pharmacokinetic, and toxicological properties using in silico tools. The compounds met key drug-likeness criteria, including Lipinski’s, Ghose’s, and Veber’s rules, supporting their potential as bioavailable anticancer agents.
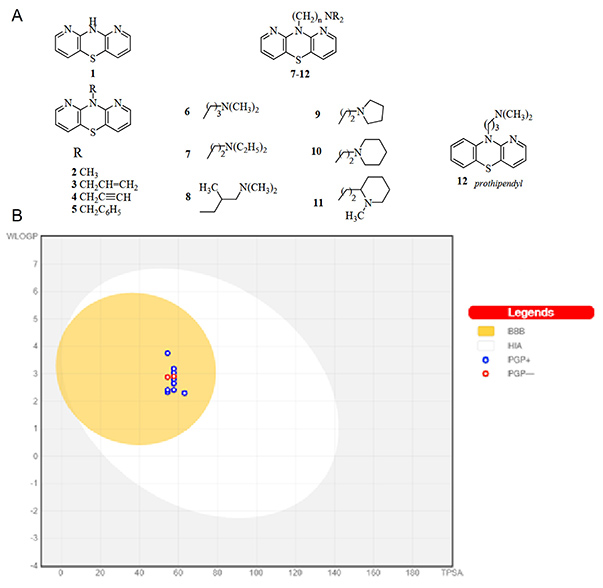
Figure 1. Graph of the dependence of lipophilicity on the polarity of the studied molecules 1–11, determined by the BOILED-Egg method. (Morak-Młodawska et al ., 2023)
Case 2: Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection
Jan et al ., 2021. doi:10.1073/pnas.2021579118
In search of COVID-19 treatments, researchers screened over 3,000 human and animal-use agents using a cell-based infection assay, identifying 15 small molecules with antiviral activity. These were further validated through enzymatic and computer modeling assays targeting viral protease and RNA polymerase. Additionally, herbal extracts showed promising results. In hamster models, mefloquine, nelfinavir, and extracts from Ganoderma lucidum , Perilla frutescens , and Mentha haplocalyx were effective against SARS-CoV-2 infection.
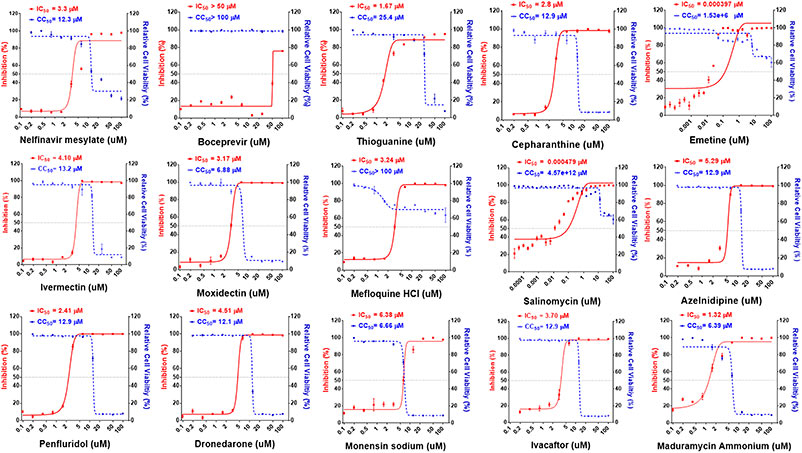
Figure 2. Dose–response relationships of 15 selected antiviral compounds. Vero E6 cells were pretreated with compounds at indicated doses followed by SARS-CoV-2 infection for 48 h. The percentage of viral titer determined by antinucleocapsid antibody after drug treatment (red) and cell viability (blue) were measured and expressed as mean ± SD of at least three independent experiments. (Jan et al ., 2021)
What Clients Say About Our Drug Analysis Expertise
“Creative BioMart's pharmacokinetics team supported our early-stage CNS compound with a full ADME package, including logD, pKa, and plasma stability assessments. Their rapid turnaround and detailed metabolic profiling helped us make a go/no-go decision before animal studies. The quality of their LC-MS/MS data was on par with top-tier CROs we've used previously, but with far more flexible communication.”
— Senior Scientist, Drug Metabolism & Pharmacokinetics | Mid-Sized Biotech Company
“We engaged Creative BioMart for CYP450 inhibition and induction analysis on a first-in-class kinase inhibitor. Their team not only identified key metabolic liabilities but also helped us understand species-specific differences using both rodent and human microsomes. Their interpretive report gave our regulatory group the confidence to advance into IND-enabling studies.”
— R&D Director | Global Pharmaceutical Company
“We needed to quantify several active components in a complex botanical extract intended for a traditional medicine trial. Creative BioMart established and validated a robust multi-component LC-MS/MS method that met both our regulatory and pharmacological endpoints.”
— Principal Investigator | Academic Research Institute
“Creative BioMart supported our bioequivalence study with full bioanalytical method development, validation, and clinical sample analysis. Their team managed over 400 plasma samples with high precision and consistency. The results not only supported our NDA submission but also helped us navigate a key discussion with regulatory reviewers.”
— Clinical Pharmacology Manager | Generic Drug Manufacturer
FAQs on Our Drug Analysis Service
-
Q: What types of compounds can you analyze under your drug analysis service?
A: We provide comprehensive analytical services for pre-clinical drug candidates, including synthetic pharmaceuticals and natural products. Whether you're assessing small molecules, herbal extracts, or active pharmaceutical ingredients (APIs), we have the tools and expertise to deliver precise data for your development stage. -
Q: What pharmacokinetics and bioanalysis services do you offer?
A: We offer both in vitro ADME studies and in vivo pharmacokinetic evaluations. This includes analysis of drug absorption, distribution, metabolism, and elimination, bioavailability studies, CYP450 enzyme interactions, metabolite identification, and bioequivalence assessments in rodent models. -
Q: Can you identify and quantify impurities in my drug compound?
A: Yes. Our pre-clinical pharmaceutical property analysis includes impurity profiling, separation and structural elucidation of degradation products, and quantification against established impurity thresholds. We also perform stress, accelerated, and long-term stability testing to support impurity evaluation under various storage conditions. -
Q: How do you ensure the accuracy and reliability of your analysis results?
A: Our analyses are conducted using state-of-the-art instrumentation such as UPLC-MS/MS, NMR (Bruker 400/600M), HRMS, FTIR, and GC-MS/MS. Each method is rigorously validated, and all procedures adhere to industry-standard protocols to ensure high accuracy, reproducibility, and regulatory compliance. -
Q: Do you provide method development and validation services?
A: Absolutely. We specialize in developing, validating, and implementing analytical methods for bioanalysis, genotoxic impurities, residual solvents, and multi-component natural products. Our validated methods ensure compliance with regulatory standards and are tailored to your specific sample type and research goals. -
Q: How do you handle complex natural product samples?
A: Our Natural Product Analysis Service is designed to handle the complexity of plant-derived compounds. We perform component identification, metabolite profiling, quantification, and metabolomics studies using HRMS and UPLC-MS/MS technologies. Whether it's a crude extract or a refined compound, we can provide detailed compositional and pharmacokinetic data. -
Q: Can I request a custom analysis package tailored to my project?
A: Yes. We understand that no two projects are alike. We work closely with our clients to design customized analysis packages based on your research objectives, regulatory needs, and timeline. Our flexible service model ensures your compound is evaluated under the most relevant conditions.
Resources
Related Services
References:
- Jan JT, Cheng TJR, Juang YP, et al . Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection. Proc Natl Acad Sci USA . 2021;118(5):e2021579118. doi:10.1073/pnas.2021579118
- Morak-Młodawska B, Jeleń M, Martula E, Korlacki R. Study of lipophilicity and ADME properties of 1,9-diazaphenothiazines with anticancer action. IJMS . 2023;24(8):6970. doi:10.3390/ijms24086970
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

