Structural Finding for Us to Better Understand the Role of MITF
Microphthalmia associated transcription factor (MITF) works as a master regulator in cells which produce melanocytes. It is crucial for the development, survival and differentiation of melanocytes. It also plays important roles in other cell type.
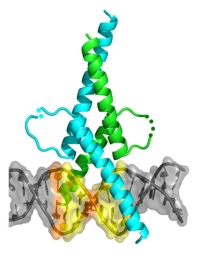
This time, scientists resolved the structure of MITF in the development of melanoma by combining structural and functional analyses. Through the study, they found MITF has unusual and different structure features comparing to similar transcription factors.
The structure shows a very unusual 3 residue insertion in the leucine zipper register resulting in a kink in one of the zipper helices. This insertion limits MITF’s ability to dimerise only with those transcription factors that also have this unusual insert. DNA binding is also shown to be very specific and mutants show an increase in non-specific DNA binding, illustrating how important this delicate balance between specific and non-specific binding is for MITF’s role as a master regulator in the development, survival and differentiation of melanocytes.
With the finding, scientists got better understanding in the function of MITF. In next step, scientists will evaluate related studies targeting on the factor. The finding also offers them a clue to explore the whole protein and its ligands for therapeutic purposes, for example.
Creative BioMart offers MITF-related products and service. Come to learn more…
Article Link: Structural Finding for Us to Better Understand the Role of MITF

