Integrated Drug Development & Manufacturing Bioprocess Development
The Role of Integrated Bioprocess Development in Drug Manufacturing
Integrated drug development and manufacturing bioprocess development is a critical and multifaceted field that bridges the gap between the discovery of new therapeutic agents and their large-scale production for clinical use. This integrated approach ensures that the bioprocesses used to manufacture drugs are optimized for efficiency, scalability, and regulatory compliance from the earliest stages of development.
Key Components
- Target Identification and Validation : This involves identifying biological targets that play a critical role in disease pathways and validating their potential for therapeutic intervention.
- Bioprocess Development : This includes optimizing cell lines, culture conditions, and purification processes to maximize yield and quality.
- Regulatory Compliance : Ensuring that bioprocesses meet stringent regulatory standards, such as those set by the FDA and EMA, is crucial. This includes validation of manufacturing processes and quality control measures.
- Technological Innovation : The use of advanced technologies, such as high-precision tunable laser spectroscopy (HPTLS) for real-time monitoring and control of bioprocesses, enhances accuracy and efficiency.
With extensive experience in mammalian cell culture systems and biologics production, Creative BioMart offers an integrated, one-stop solution for your drug development and manufacturing needs—from cell line development to quality-controlled production.
Our End-to-End Bioprocess Development and Manufacturing Services
At Creative BioMart, we provide comprehensive bioprocess development services tailored for biopharmaceutical companies seeking end-to-end support. Our integrated approach combines robust scientific expertise, cutting-edge technology platforms, and stringent quality systems to streamline your journey from DNA to clinical-grade material.
We specialize in:
- Stable cell line development using CHO, NS0, and HEK293 systems
- Scalable upstream and downstream process development
- Single-use bioreactor systems for flexible and cost-effective production
- Integrated QA/QC systems guided by Quality by Design (QbD) principles
|
 |
 |
|
|
Integrated Quality Assurance & Quality Control (QA/QC)
|
 |
Why Our Integrated Approach Enhances Drug Development Efficiency
- Comprehensive Expertise : From cell line development to manufacturing, all under one roof.
- Speed & Flexibility : Rapid timelines enabled by DOE and single-use bioreactor systems.
- Regulatory Readiness : QbD-driven processes built to meet global regulatory standards.
- Cost Efficiency : Optimized workflows reduce development time and production cost.
- Dedicated Support : Cross-functional teams ensure smooth tech transfer and clear communication at every step.
- Seamless Integration : Harmonized development and manufacturing workflows ensure continuity, reducing risk during scale-up and tech transfer.
Case Studies on Integrated Drug Development and Bioprocessing Success
* NOTE: We prioritize confidentiality to safeguard our clients’ technology and intellectual property. As an alternative, we present selected published research articles as representative case studies. For details on the assay services and products used in these studies, please refer to the relevant sections of the cited literature.
Case 1: High-performance recombinant protein production with Escherichia coli in continuously operated cascades of stirred-tank reactors
Schmideder and Weuster-Botz, 2017. doi:10.1007/s10295-017-1927-y
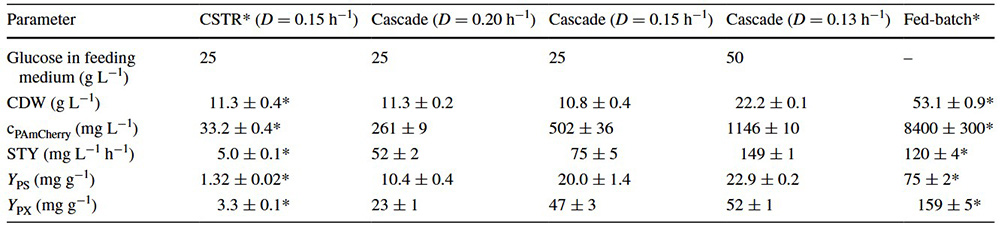
This study developed a continuous bioprocess using a cascade of two stirred-tank reactors to improve intracellular production of photoactivatable mCherry protein in E. coli . Biomass formation and protein expression were spatially separated between the reactors. Using miniaturized stirred-tank cascades, the researchers enabled 24-fold parallel testing and direct scalability. The process achieved high protein concentration (1.15 g/L) and significantly outperformed traditional chemostats and optimized fed-batch processes in space-time yield (149 mg/L/h). The findings highlight continuous cascade systems as a promising, efficient alternative to fed-batch production and show that miniaturized reactors can accelerate process development while reducing costs.
Table 1. Comparison of bioprocesses for the production of PAm-Cherry with E. coli in continuous stirred-tank reactors (CSTR), in continuously operated cascades of two stirred-tank reactors operated at different dilution rates (D), and an optimized fed-batch processes regarding cell dry weight (CDW), PAm-Cherry concentration (cPAm-Cherry), space–time yield (STY), product selectivity (YPS), and product yield coefficient (YPX). (Schmideder and Weuster-Botz, 2017)
Case 2: Monoclonal antibody production with an unnatural amino acid in Pichia pastoris
Tir et al. , 2022. doi:10.1186/s12934-022-01882-6
This study used Pichia pastoris to produce trastuzumab IgG containing the unnatural amino acid p-azido-l-phenylalanine (pAzF) through genetic code expansion. By modifying secretion signals, overexpressing ER chaperones Kar2p and Lhs1p, and knocking out vacuolar degradation in Fab-producing strains, protein yields improved significantly. Fed-batch bioreactor cultivation boosted productivity up to 238 mg/L for IgG and 15 mg/L for IgGpAzF—20–50 times higher than initial screenings. Site-specific incorporation of pAzF was confirmed by mass spectrometry. The work demonstrates cost-effective, lab-scale production of modified antibodies and supports the use of P. pastoris for developing site-specific conjugates in antibody engineering.
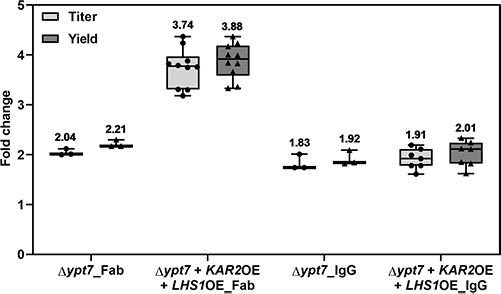
Figure 1.Influence of YPT7 disruption and KAR2 and LHS1 co-overexpression on Fab and IgG production. (Tir et al. , 2022)
What Clients Say About Our Integrated Bioprocess Solutions
“Creative BioMart accelerated our cell line development with impressive speed and consistency. Their DOE approach helped us achieve expression levels we hadn’t seen in previous attempts.”
— Director of Cell Line Engineering | Biopharma Company
“The transition from lab to pilot scale was seamless thanks to their single-use bioreactor system and robust QA oversight. Highly recommend for early-phase biologics.”
— VP of Manufacturing | Oncology-Focused Biotech
“We appreciated the integrated project management and regulatory focus throughout the process. Creative BioMart’s QbD mindset really shows.”
— Head of CMC | Global Pharma Partner
“Thanks to Creative BioMart’s rapid clone selection and scale-up process, we shaved months off our development timeline without compromising quality.”
— Senior Scientist | Preclinical Biotech Startup
FAQs on Integrated Bioprocess Development and Manufacturing
-
Q: What types of cell lines do you support for development?
A: We work extensively with CHO, NS0, and HEK293 cells, and can accommodate other mammalian systems upon request. -
Q: How fast can you deliver a stable cell line?
A: With our DOE-based screening and automation, we typically deliver high-yield stable lines in 6–8 weeks. -
Q: Do you offer GMP manufacturing support?
A: Yes. Our process development is fully scalable and can be seamlessly transitioned to GMP production upon request. -
Q: How do you ensure quality during development?
A: Our QA/QC systems follow SOPs rooted in QbD principles, with full traceability, documentation, and analytical testing.
Resources
Related Services
- GMP Stable Cell Line Development Platform
- Cell Line Development and Membrane Preparation
- Stable Cell Line Services
- Human Cell Line Activation Test (h-CLAT)
- Downstream Processing Development
- Quality Control
- Protein Expression and Purification Services
- Drug Discovery Screening
- High-Throughput Screening
Related Products
References:
- Schmideder A, Weuster-Botz D. High-performance recombinant protein production with Escherichia coli in continuously operated cascades of stirred-tank reactors. Journal of Industrial Microbiology and Biotechnology . 2017;44(7):1021-1029. doi:10.1007/s10295-017-1927-y
- Tir N, Heistinger L, Grünwald-Gruber C, et al. From strain engineering to process development: monoclonal antibody production with an unnatural amino acid in Pichia pastoris . Microb Cell Fact . 2022;21(1):157. doi:10.1186/s12934-022-01882-6
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

