Principle and Protocol of Biuret Method
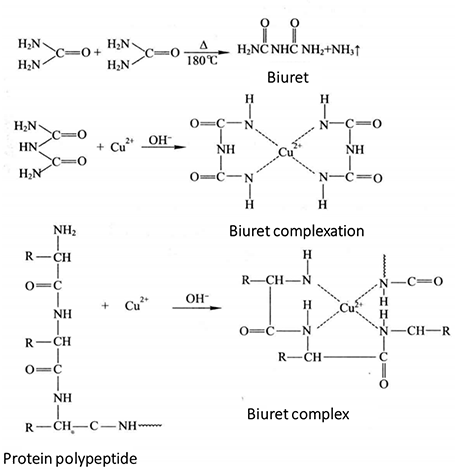
When the urea is heated to about 180 °C, one molecule of ammonia is released from two molecules of urea to form biuret (NH2-CO-NH-CO-NH2). Biuret combines with Cu2+ in alkaline solution to form purple complex, which is called biuret reaction.
The protein contains more than two peptide bonds, so there is a biuret reaction. In alkaline solution, protein forms a purple red complex with Cu2+. Its color is in direct proportion to the concentration of protein, but has nothing to do with the relative molecular weight of protein and amino acid composition. Therefore, the protein content can be determined by colorimetry. In addition to -CONH-, this reaction is also found in -CONH2-, -CH2-, NH2-, -CS-CS-NH2 and other groups. Under certain conditions, the solution of the unknown sample reacts with the standard protein solution at the same time, and the color is compared at 540-560 nm. The unknown protein concentration can be calculated from the standard curve of the standard protein.
1. Main Instruments and Equipment
Micropipette, ultraviolet visible spectrophotometer, quartz cuvette, balance.
2. Experimental Materials
Protein samples extracted by various methods.
3. Main reagents
Sample solution*1
Protein standard solution (10 mg/mL): accurately weigh 50 mg of bovine serum albumin and prepare 10 mg/mL with 5 mL of sample solution.
Biuret reagent: weigh 1.50 g of copper sulfate (CuSO4 · 5H2O) and 6.0 g of potassium sodium tartrate (KNaC4H4O6 · 4H2O), dissolve them with 500 mL of water, add 300 mL of 10 % NaOH solution under stirring, dilute them with water to 1L, and store them in a plastic bottle*2 (or a bottle coated with paraffin on the inner wall).
1. Standard Curve Method
(1) Take 7 tubes and add reagents according to Table 2-1-3
Table 2-1-3 Preparation of Protein Standard Curve*3
| Reagent | Test tube number | ||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Standard solution /mL | - | 0.1 | 0.2 | 0.4 | 0.6 | 0.8 | 1 |
| Sample solution /mL | 0.1 | 0.9 | 0.8 | 0.6 | 0.4 | 0.2 | - |
| Protein concentration mass /(mg/mL) | - | 1 | 2 | 4 | 6 | 8 | 10 |
Figure 2-1-3 Standard Curve of Biuret Method
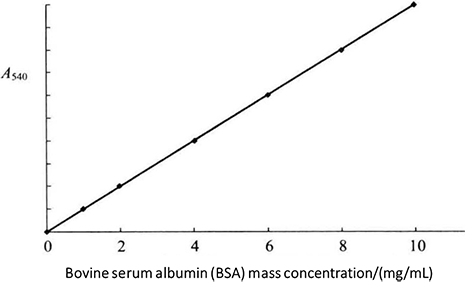
(2) Add 4mL biuret reagent into each tube. After fully shaking, place it at room temperature (20-25°C) for 30min, and conduct colorimetric measurement at 540mm. Use the first tube without protein solution as the blank control solution. The standard curve is drawn with the protein content as the abscissa and the absorbance value as the ordinate (Figure 2-1-3).
2. Determination of Protein Content in Samples
Take 1mL of protein sample to be tested, add 4.0mL of biuret reagent, mix well and place for 30min, then measure the absorbance value by colorimetry under the wavelength of 540mm, and find out the mass concentration of protein according to the standard curve. Note that the mass concentration of the sample shall not exceed 10mg/mL *4. If it exceeds this mass concentration, the corresponding multiple shall be diluted, and the measured result shall be multiplied by the dilution multiple to obtain the true mass concentration of the protein sample to be tested.
1. It must be determined by colorimetry within 30min after color development. The time from color development to color comparison of each tube shall be as consistent as possible.
2. When a large amount of fatty substances exist at the same time, a turbid reaction mixture will be produced. At this time, ethanol or petroleum ether can be used to clarify the solution, centrifugate it, and then take the supernatant for determination.
* 1 This can be distilled water, protein buffer solution or salt solution. Depending on the sample dissolution solution.
* 2 This reagent can be stored for a long time. If a black precipitate appears in the storage bottle, it needs to be reconstituted.
* 3 Standard solution preparation.
*4 In a certain concentration range, the absorbance is proportional to the protein concentration.

