Protein Engineering (Mutant Libraries)
Creative BioMart offers cutting-edge Protein Engineering Services, specializing in the design and construction of Mutant Libraries for advanced research and industrial applications. Leveraging our robust de novo gene synthesis platform, we deliver custom site-directed, scanning, randomized, degenerated, and truncation libraries without inflating your R&D budget. These libraries enable high-throughput screening of protein function, structural insights, and identification of critical regulatory regions—all with unparalleled accuracy, diversity, and speed.
Background: In Vitro Molecular Optimization of Proteins
Types of Mutant Libraries
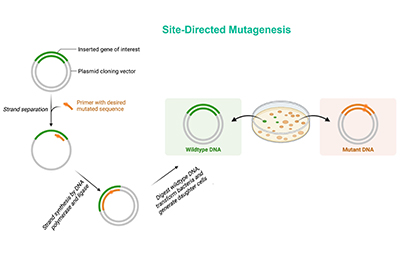
- Site-Directed Mutagenesis Libraries
These libraries involve the systematic substitution of specific amino acids with other amino acids. This method is useful for studying the functional and structural roles of individual residues in a protein.
- Scanning Point Mutation Libraries
This method systematically replaces each amino acid in a protein with all 20 amino acids simultaneously. This provides a detailed profile of each position in the protein and helps identify critical residues for function and stability.
- Randomized and Degenerated Libraries
These libraries introduce random mutations across the entire protein sequence or specific regions. This is often achieved using degenerate codons, which can encode multiple amino acids. This method is useful for exploring a large sequence space and identifying beneficial mutations.
- Precision Mutant Libraries
Precision mutant libraries are high-quality and diverse synthetic libraries that contain only desired mutants. These libraries are created using advanced DNA synthesis techniques and automated platforms, allowing for the incorporation of individually specified nucleotides. This approach significantly reduces screening efforts and increases the likelihood of finding beneficial variants.

Applications of Mutant Libraries
- Enzyme Engineering
Mutant libraries are used to optimize enzymes for industrial applications, such as improving catalytic efficiency, thermal stability, and substrate specificity. For example, directed evolution techniques can generate random mutations in a gene, followed by rapid screening to identify variants with desired properties.
- Antibody Engineering
Mutant libraries are used to improve the binding affinity and specificity of antibodies. This is crucial for developing therapeutic antibodies with high efficacy and reduced off-target effects.
- Protein Folding and Solubility
Mutant libraries can help identify mutations that improve protein folding and solubility, which is important for both research and industrial applications.
- Phosphorylation Sites and Protein-Protein Interactions
Mutant libraries can be used to study phosphorylation sites and protein-protein interactions, providing insights into cellular signaling pathways and potential therapeutic targets.
Advantages of Mutant Libraries
- Comprehensive Exploration : Mutant libraries allow for the exploration of a large sequence space, increasing the chances of finding beneficial mutations.
- Targeted Screening : Precision mutant libraries enable targeted screening, reducing the number of clones that need to be analyzed.
- Improved Variants : By systematically introducing mutations, researchers can identify variants with enhanced properties, leading to improved protein performance
Modern protein engineering relies on precise control over amino acid sequences to unlock new or improved protein functions. In vitro molecular optimization is a powerful strategy for generating mutant variants to explore structure-function relationships, identify essential residues, and engineer regulatory features. At Creative BioMart, we apply this technique to build highly versatile mutant protein libraries, enabling systematic studies in protein performance, regulation, and domain mapping.
Service Overview: Comprehensive Mutant Library Construction
Protein Engineering Service Procedure
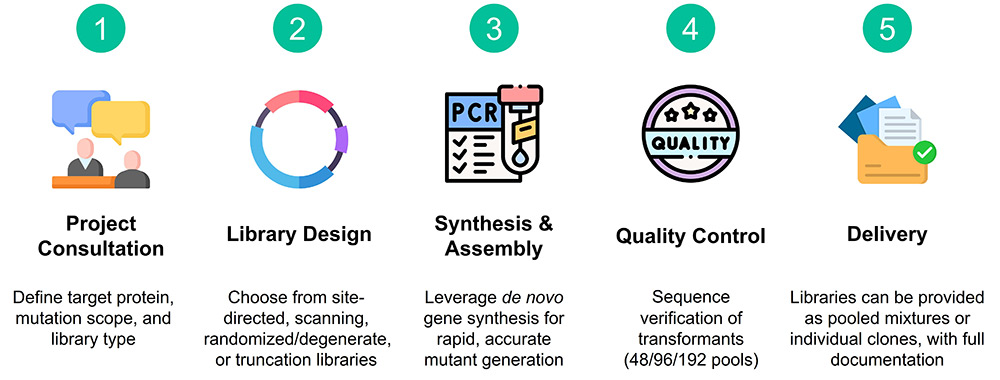
Protein Mutant Library Service Details
|
Service |
 |
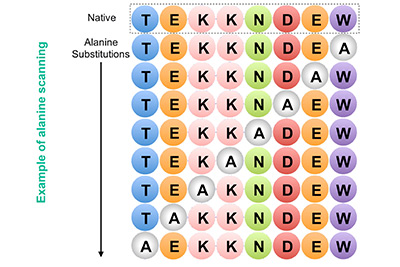 Scanning Point Mutation Libraries (Sequential Permutation Libraries) |
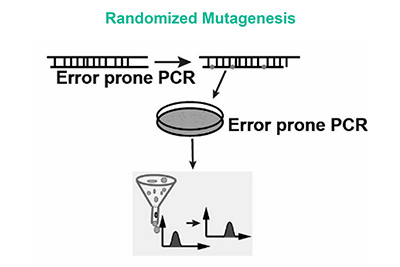 |
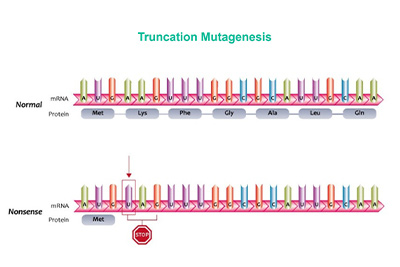 Truncation Libraries |
|
Details |
|
|
|
|
Why Choose Our Mutant Library-Based Protein Engineering Solutions
- Unmatched Library Diversity : Comprehensive mutant coverage, from single-residue swaps to randomized domains.
- High Integrity DNA : >90% sequence accuracy with minimal off-target mutations.
- Flexible Formats : Choose pooled libraries or individual transformants tailored to your screening strategy.
- Advanced Design, Simplified Execution : Our synthesis-driven approach removes bottlenecks in mutant generation.
- Application-Driven Engineering : Optimize solubility, identify functional cores, or map epitopes with ease.
- Expert Support : Work with protein engineering specialists throughout your project lifecycle.
Case Studies in Protein Engineering Using Mutant Libraries
* NOTE: We prioritize confidentiality to safeguard our clients’ technology and intellectual property. As an alternative, we present selected published research articles as representative case studies. For details on the assay services and products used in these studies, please refer to the relevant sections of the cited literature.
Case 1: Protein engineering with mutant libraries to enhance anti-PD-1 nanobody affinity
Mirzaei et al. , 2025. doi:10.1007/s12033-024-01162-1
To develop more effective PD-1 inhibitors, researchers engineered nanobodies using CDR grafting from the antibody tislelizumab into a nanobody framework. To enhance binding affinity, site-directed mutagenesis was used to create two mutant variants with substitutions (Tyr97Arg and Tyr102Arg) in the VHH-CDR3 region. These mutations significantly improved nanobody binding to PD-1, as confirmed by Dot blot, Western blot, and ELISA. This study highlights how protein engineering and mutant libraries can be used to optimize therapeutic nanobodies, offering a promising alternative to traditional antibodies in immunotherapy.
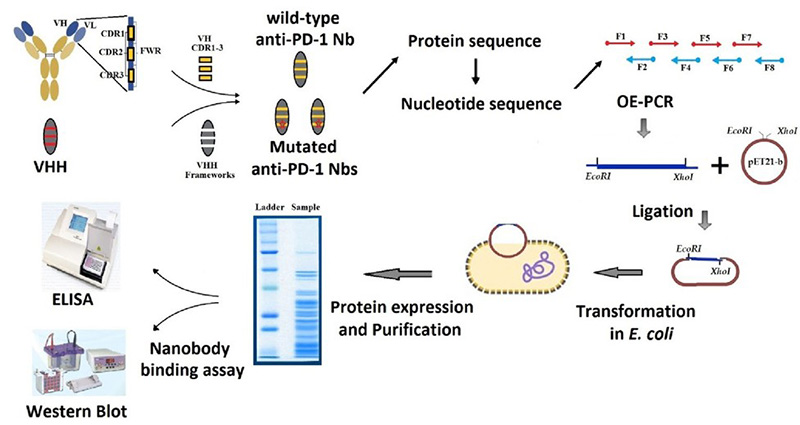
Figure 1. Distribution of detection of preservatives . A Distribution of detecting dehydroacetic acid . B Distribution of detecting benzoic acid . C Distribution of detecting of ethylparaben . D Distribution of detecting of sorbic acid . (Mirzaei et al., 2025)
Case 2: Protein engineering with mutant libraries reveals key residue in novel esterase
Lee et al ., 2021. doi:10.1016/j.ijbiomac.2021.08.224
A novel esterase gene, est3S , was identified from a metagenomic library of cow rumen microbes and characterized through protein engineering. The 342-amino-acid enzyme showed unique properties—activity at pH 7.0 and 40 °C, partial solvent resistance, and strong degradation of pesticides like chlorpyrifos and methyl parathion. Using site-directed mutagenesis, the Ser132 residue was replaced (Ser132Ala), resulting in complete loss of activity, confirming its critical role. This study highlights how mutant libraries can elucidate structure–function relationships, enabling the identification of essential catalytic residues and guiding future engineering of enzymes for bioremediation and industrial use.
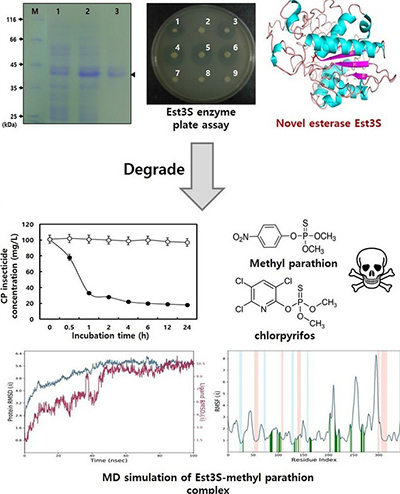
Figure 3. Mining of a novel esterase ( est3S ) gene from a cow rumen metagenomic library with organosphosphorus insecticides degrading capability: Catalytic insights by site directed mutations, docking, and molecular dynamic simulations. (Lee et al. , 2021)
Customer Testimonials: Success Stories with Mutant Library Protein Engineering
"We collaborated with Creative BioMart to develop a site-directed mutagenesis library targeting an allosteric site on one of our GPCR candidates. The turnaround time was excellent, and the quality of the constructs exceeded our expectations—over 95% of clones were mutation-accurate. This allowed our team to quickly pinpoint key residues for ligand modulation, significantly accelerating our hit-to-lead process."
— R&D Director | Mid-Sized Pharmaceutical Company
"Our lab needed a truncation library to map the minimal DNA-binding domain of a novel transcription factor we identified in plant systems. Creative BioMart’s team was responsive, technically thorough, and delivered a library with impressive diversity (~35,000 variants). Every construct maintained the correct reading frame, saving us from tedious downstream validation. Their service made a complex project straightforward."
— Principal Investigator | Academic Research Institution
"We commissioned a degenerate library covering the active site of an enzyme we’re optimizing for industrial catalysis. Creative BioMart’s synthesis-based approach allowed us to precisely control the mutation frequency, which was critical for our structure-function mapping. The sequence-verified transformant pool gave us a reliable starting point, and we’ve already identified several promising variants with >5x catalytic efficiency."
— Protein Engineering Lead | Biotechnology Startup
"Creative BioMart built a scanning point mutation library for our work on antibody affinity maturation. Unlike typical alanine scans, their method gave us comprehensive substitution data at each position, which directly led to the identification of a previously unknown stabilizing mutation."
— Senior Scientist | Government-Funded Biomedical Research Center
FAQs on Protein Engineering and Mutant Library Services
-
Q: What types of mutant libraries do you offer?
A: We offer a full spectrum of mutant libraries, including:- Site-directed mutagenesis libraries (single-residue substitutions)
- Scanning point mutation libraries (each residue replaced with all 20 amino acids)
- Randomized and degenerated libraries (precise or broad-spectrum random mutations)
- Truncation libraries (5’ and 3’ incremental gene trimming around a core region)
-
Q: How accurate are the mutant libraries you construct?
A: Thanks to our advanced de novo gene synthesis and quality control systems, our libraries maintain >90% sequence integrity, depending on gene length. For sequence-verified transformant pools (48, 96, or 192 clones), each clone is individually verified to ensure high accuracy and functional relevance. -
Q: Can you control the mutation frequency in randomized libraries?
A: Absolutely. We offer precise control over mutation frequency, from 1 to 20 mutations per kilobase, using degenerate oligonucleotide synthesis. This allows you to fine-tune the balance between diversity and function. -
Q: What formats can I receive the library in?
A: We offer maximum flexibility: libraries can be delivered as pooled mixtures or as individual clones. Depending on your screening workflow or throughput preferences, we tailor the format to best suit your needs. -
Q: What are the typical applications for your mutant libraries?
A: Our libraries are used widely in:- Protein function and structure analysis
- Active site and regulatory sequence mapping
- Affinity maturation
- Solubility and stability optimization
- Epitope mapping and inhibitor screening
-
Q: What makes your mutant library services different?
A: We combine state-of-the-art de novo gene synthesis, sequence-verified delivery, and custom mutation control into a seamless, cost-effective workflow. Our ability to synthesize complex libraries without dramatically increasing costs gives you access to high-quality, diverse libraries with unmatched reliability.
Resources
Related Services
References:
- Mirzaei M, Mirhoseini S, Heidari MM, Khatami M. Design and production of a novel anti-PD-1 nanobody by CDR grafting and site-directed mutagenesis approach. Mol Biotechnol. 2025;67(5):1843-1851. doi:10.1007/s12033-024-01162-1
- Fürst MJLJ, Martin C, Lončar N, Fraaije MW. Experimental protocols for generating focused mutant libraries and screening for thermostable proteins. In: Methods in Enzymology. Vol 608. Elsevier; 2018:151-187. doi:10.1016/bs.mie.2018.04.007
- Xu Y, Wu Y, Lv X, et al. Design and construction of novel biocatalyst for bioprocessing: Recent advances and future outlook. Bioresource Technology. 2021;332:125071. doi:10.1016/j.biortech.2021.125071
- Campbell NA. Biology. 2. ed. Benjamin/Cummings Pub. Co; 1990.
- Lee HY, Cho DY, Ahmad I, et al. Mining of a novel esterase (est3s) gene from a cow rumen metagenomic library with organosphosphorus insecticides degrading capability: Catalytic insights by site directed mutations, docking, and molecular dynamic simulations. International Journal of Biological Macromolecules. 2021;190:441-455. doi:10.1016/j.ijbiomac.2021.08.224
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

