Cell-free Display Platform
Creative BioMart provides comprehensive Cell-free Display Platform Services for protein engineering and directed evolution. Free from the constraints of living cells, our in vitro systems—including ribosome display, mRNA display, covalent DNA display, and in vitro compartmentalization (IVC)—enable screening of ultra-large variant libraries (up to 10¹⁴ ). These systems allow precise control over reaction conditions and facilitate the incorporation of non-natural amino acids or nucleotides. Our expertise in optimizing display conditions, enriching desired variants, and characterizing functional proteins ensures an efficient and flexible approach for evolving high-performance molecules.
Background: Understanding Cell-free Display
Cell-free display platforms represent a powerful evolution of traditional display technologies by eliminating the need for living cells. In contrast to in vivo systems, these in vitro approaches bypass transformation efficiency limitations, allowing the creation and selection of libraries several orders of magnitude larger. Moreover, the cell-free environment permits chemical modification of nucleic acid and amino acid components, expanding the functional diversity of screened proteins.
Cell-free display and selection systems generally follow three key steps: library construction, enrichment of desired variants through biopanning, and functional screening and characterization of selected candidates. A crucial aspect of these methods is maintaining a reliable genotype–phenotype linkage to connect each protein with its encoding gene. For example, in ribosome display, this linkage is maintained via stable protein–ribosome–mRNA (PRM) complexes, while in mRNA display, it is achieved by attaching puromycin to the mRNA’s 3′ end, ensuring that the translated protein remains covalently linked to its genetic information for efficient selection.
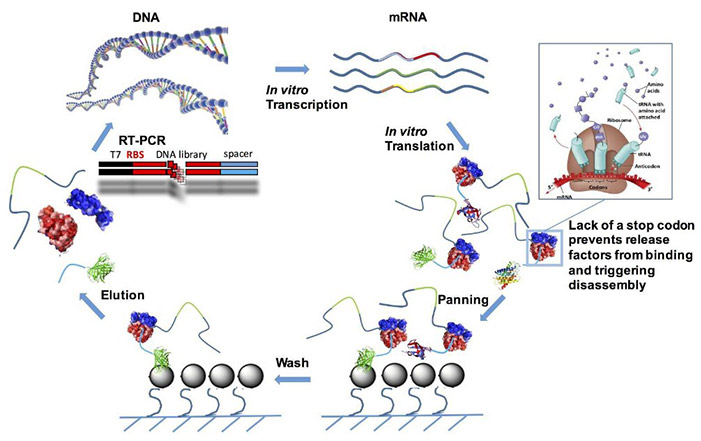
Figure 1. Scheme of ribosome display selection cycle. (Galán et al., 2016)
Cell extracts from E. coli, wheat germ, or rabbit reticulocytes supply the translational machinery needed for protein synthesis, providing an open and customizable platform. This makes cell-free display ideal for evolving proteins with improved affinity, stability, specificity, or catalytic activity, with broad applications in therapeutic antibody development, enzyme engineering, and diagnostics.
What Our Special Cell-free Display Platform Deliver
Creative BioMart provides end-to-end solutions in cell-free display for directed evolution and protein optimization. Our services include:
- Design and implementation of custom cell-free display systems tailored to your molecular target
- Library construction and optimization for ultra-high diversity (up to 10¹⁴ variants)
- Variant enrichment through iterative biopanning cycles to identify desired properties
- Functional screening and characterization using customized biochemical or binding assays
- Incorporation of unnatural amino acids or non-natural nucleotides for specialized applications
- Comprehensive data reporting and post-selection analysis to support downstream development
Service Workflow

Service Details
|
Parameter |
Details |
|---|---|
|
Library Diversity |
Up to 10¹⁴ unique variants. |
|
Available Platforms |
Ribosome display, mRNA display, covalent/non-covalent DNA display, and in vitro compartmentalization (IVC). |
|
Genotype–Phenotype Linkage |
Ribosome–mRNA complex or puromycin linkage. |
|
Source Extracts |
E. coli, wheat germ, rabbit reticulocyte, or custom hybrid systems. |
|
Screening Methods |
Biopanning, fluorescence-based assays, and affinity chromatography. |
|
Applications |
Antibody discovery, enzyme evolution, peptide optimization, diagnostic biomolecule development. |
Why Choose Us
- Unmatched Library Diversity: Display and screen up to 10¹⁴ variants for unparalleled selection depth.
- Flexible Platform Options: Choose from multiple cell-free systems optimized for your target molecule.
- Precise Reaction Control: Fully defined in vitro environment allows fine-tuning of translation and selection conditions.
- Expanded Chemical Space: Incorporate unnatural amino acids and nucleotides to enhance protein functionality.
- Expertise in High-throughput Screening: Seamless integration with advanced biopanning and FACS technologies.
- Comprehensive One-stop Service: From design to functional characterization, all handled by our experienced protein engineers.
Case Studies of Protein Engineering Using a Cell-free Display Platform
Case 1: Efficient protein selection based on ribosome display system with purified components
Ohashi et al., 2007. doi:10.1016/j.bbrc.2006.11.017
Using the PURE (Protein synthesis Using Recombinant Elements) system, researchers developed a highly efficient and controllable ribosome display method for selecting functional proteins. Composed entirely of purified E. coli transcription–translation components, the PURE system enabled precise optimization and stable formation of mRNA–ribosome–polypeptide complexes, a key factor for successful display. These improvements led to a dramatic increase in mRNA recovery and achieved approximately 12,000-fold enrichment of single-chain antibody (scFv) cDNA in just one selection round. Remarkably, specific scFv selection from a 1:10¹⁰ mRNA mixture was completed in three rounds, confirming the PURE system’s reliability and reproducibility for ribosome display applications.
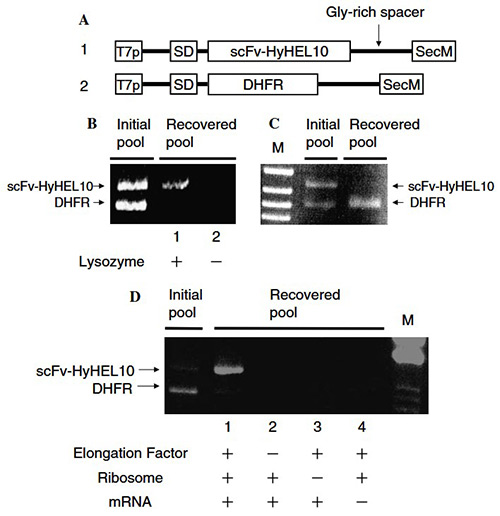
Figure 2. Construction and validation of the pure ribosome display (PRD) system. (A) DNA constructs of scFv-HyHEL10 and DHFR under T7 promoter with a Gly-rich linker and SecM sequence. (B,C) Affinity selection of scFv for lysozyme and DHFR for MTX from mixed mRNA pools, analyzed by RT-PCR. (D) Verification of mRNA–ribosome–polypeptide complex formation by testing the effect of removing translation components. (Ohashi et al., 2007)
Case 2: The use of mRNA display to select high-affinity protein-binding peptides
Wilson et al., 2001. doi:10.1073/pnas.061028198
This study demonstrates the use of mRNA display, an in vitro selection technique, to identify high-affinity peptide aptamers for streptavidin. Using an exceptionally large library of ~10¹³ random peptides, researchers isolated 20 distinct aptamers with dissociation constants as low as 5 nM—comparable to monoclonal antibody–antigen interactions. Unlike conventional systems, these aptamers required no disulfide bridges or engineered scaffolds. They exhibited binding affinities 1,000–10,000 times stronger than those produced via phage display from smaller libraries. The presence of an HPQ consensus motif among selected peptides highlights how ultrahigh-diversity mRNA display can generate potent, specific binders from unconstrained peptide libraries.
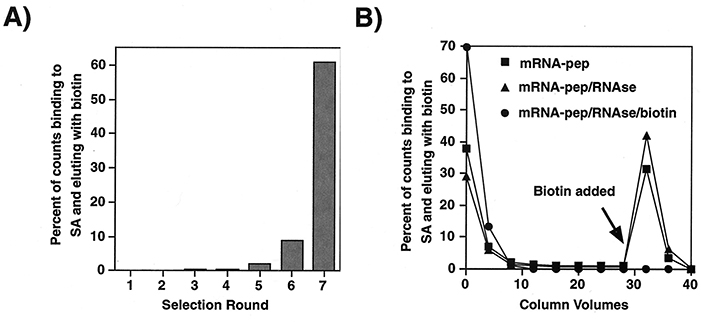
Figure 3. Selection progress and results. (A) Fraction of displayed peptides bound to streptavidin and eluted with biotin in each selection round. (B) Elution profile of peptides from the seventh-round library, comparing intact mRNA–peptide complexes, RNase-treated samples, and biotin-blocked controls. (Wilson et al., 2001)
What Our Clients Say About Our Cell-free Display Platform
“We collaborated with Creative BioMart to improve the affinity of an antibody fragment targeting a difficult-to-express membrane protein. Their cell-free ribosome display platform allowed us to screen an extraordinarily large library—something we could never have achieved with phage display. Within weeks, we identified clones with over 25-fold higher binding affinity. The team’s technical expertise and transparent communication made the entire process efficient and scientifically rewarding.”
— Senior Scientist, Antibody Discovery | Global Biopharmaceutical Company
“Our R&D group approached Creative BioMart to enhance the thermal stability of a proprietary industrial enzyme. Their in vitro compartmentalization (IVC) approach was impressively flexible, enabling the incorporation of unnatural amino acids into our variants. The final mutants not only exhibited a 3× increase in activity retention at high temperature but also maintained full catalytic efficiency. The ability to fine-tune reaction conditions in a completely cell-free system gave us results that traditional directed evolution couldn’t match.”
— Head of Enzyme Engineering | Specialty Chemicals Manufacturer
“We engaged Creative BioMart’s team for rapid discovery of high-affinity peptide ligands for a diagnostic biosensor. Using their mRNA display workflow, they generated a massive peptide library and completed three selection rounds in record time. The final peptides demonstrated nanomolar affinity and excellent specificity in our surface plasmon resonance assays. The entire project—from library design to validation—was executed flawlessly, saving us months of development time.”
— Director of Molecular Diagnostics | Medical Device Company
“Our company needed to evolve a binding domain with both enhanced affinity and pH stability for a therapeutic fusion protein. Creative BioMart’s cell-free display platform provided the scale and precision required. Their scientists expertly customized a hybrid wheat germ–E. coli extract system to support our challenging construct. The resulting variants exceeded our binding and stability benchmarks, allowing us to advance to preclinical formulation faster than expected. Their collaborative spirit and deep technical know-how make them a trusted partner for complex protein projects.”
— VP of Research and Development | Mid-size Biotech Company
FAQs About Our Cell-free Display Platform
-
Q: What are the main advantages of using a cell-free display platform instead of a traditional cell-based system?
A: Cell-free display systems offer unmatched flexibility and diversity. Unlike in vivo methods, there’s no need for transformation, which means library sizes can reach up to 10¹⁴ variants, greatly expanding the sequence diversity under selection. In addition, our cell-free setup allows incorporation of unnatural amino acids and modified nucleotides, enabling evolution of proteins with novel structures and properties that are difficult—or even impossible—to achieve using cellular hosts. -
Q: What types of cell-free display technologies do you offer?
A: We provide a comprehensive suite of cell-free display systems, including ribosome display, mRNA display, covalent/non-covalent DNA display, and in vitro compartmentalization (IVC). Each method can be tailored to specific project needs—whether you’re optimizing antibody affinity, evolving enzymes for stability, or screening peptides for diagnostics. -
Q: How large can the libraries be in your cell-free display system?
A: Our cell-free systems routinely handle libraries with diversities of up to 10¹⁴ variants, far exceeding the limits of cell-based approaches. This immense diversity provides a greater chance of identifying rare, high-performance variants with superior binding, catalytic, or stability characteristics. -
Q: What kinds of projects benefit most from your cell-free display service?
A: Our platform is ideal for projects requiring high-throughput screening and rapid evolution, such as antibody or peptide affinity maturation, enzyme engineering, and discovery of binders against unstable or toxic targets. It’s also particularly useful when cell-based expression is challenging or when unnatural components are needed for protein design. -
Q: Can you help with downstream analysis and characterization?
A: Yes. Beyond the display and selection steps, our scientists provide comprehensive functional screening, biophysical characterization, and sequence verification of selected variants. We ensure every hit is fully validated, ready for downstream applications such as structural studies, biocatalysis, or therapeutic development. -
Q: How do you ensure the success of complex or customized projects?
A: Every project begins with a detailed evaluation of your target and goals. Our scientists then design a customized cell-free system—selecting the most suitable extract (E. coli, wheat germ, or hybrid system) and optimizing every supplement for best yield and fidelity. Coupled with rigorous quality control, biopanning validation, and experienced technical support, we guarantee high reliability and reproducibility from start to finish. -
Q: How long does a typical cell-free display project take?
A: Project timelines vary by complexity, but most cell-free display campaigns—covering library generation, selection rounds, and initial validation—can be completed within 6–12 weeks, significantly faster than equivalent cell-based directed evolution processes.
Other Resources
Related Services
- Protein Engineering Services
- Directed Evolution
- Protein Characterization
- Phage Display Platform
- Escherichia coli Display Platform
- Yeast Display Platform
- Special Cell-based Display Platform
- Protein Expression and Purification Services
Related Products
References:
- Galán A, Comor L, Horvatić A, et al. Library-based display technologies: where do we stand? Mol BioSyst. 2016;12(8):2342-2358. doi:10.1039/C6MB00219F
- Ohashi H, Shimizu Y, Ying BW, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochemical and Biophysical Research Communications. 2007;352(1):270-276. doi:10.1016/j.bbrc.2006.11.017
- Wilson DS, Keefe AD, Szostak JW.The use of mRNA display to select high-affinity protein-binding peptides. Proc Natl Acad Sci USA. 2001;98(7):3750-3755. doi:10.1073/pnas.061028198
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

