Escherichia coli Display Platform
The Escherichia coli ( E. coli ) display platform is a powerful and versatile system for directed evolution and protein engineering. As the most widely used Gram-negative bacterium for recombinant protein production, E. coli offers excellent genetic accessibility, rapid growth, and compatibility with high-throughput screening methods such as fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS). At Creative BioMart, we have developed optimized E. coli display strategies for surface expression, mutagenesis, and selection of proteins with improved stability, activity, or binding properties. Our one-stop services enable rapid, efficient, and customized evolution of proteins for diverse research and industrial applications.
Understanding E. coli Display Technology
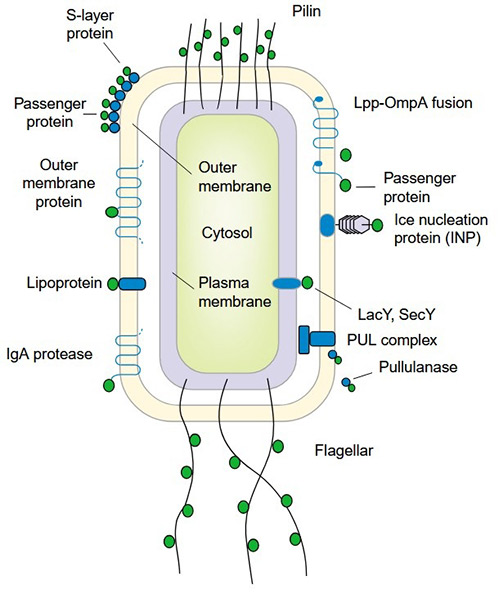
Figure 1. Cell surface display systems in Gram-negative bacteria. (Lee et al., 2003)
Since its introduction as a recombinant expression host, E. coli has become one of the most extensively employed systems for cell surface display and directed evolution. This system allows researchers to present peptides or proteins on the bacterial surface, where they can interact directly with target ligands, substrates, or receptors.
Display on the E. coli surface typically requires a carrier or anchoring motif that links the target protein to the cell envelope. Given the complexity of the Gram-negative cell wall, multiple display strategies have been developed, including:
- Insertion of target sequences into outer membrane proteins.
- Fusion to cell surface structures such as flagella.
- Attachment to the N-terminus of lipoproteins for stable surface localization.
Over the past decade, E. coli display has also been adapted for inner membrane protein evolution, opening new possibilities for engineering complex targets such as G protein-coupled receptors (GPCRs) and transporters.
Moreover, E. coli is suitable for in vivo mutagenesis to generate genetic diversity. Low-fidelity DNA polymerase mutants were introduced to promote plasmid-targeted mutagenesis. These pioneering strategies laid the groundwork for modern, continuous evolution systems that can be extended to other organisms like Saccharomyces cerevisiae.
E. coli Display Platform vs. E. coli Protein Expression System
While both platforms use E. coli as a host, they serve distinct purposes in the protein engineering workflow. The E. coli display platform is mainly designed for screening and directed evolution, enabling the discovery of superior protein variants. In contrast, the E. coli expression system focuses on efficient large-scale protein production once the best variant has been identified.
|
Category |
E. coli Display Platform |
E. coli Protein Expression System |
|---|---|---|
|
Main Purpose |
Directed evolution and selection of optimized protein variants |
High-yield recombinant protein production |
|
Expression Location |
Protein displayed on the cell surface |
Protein produced inside the cell or secreted |
|
Key Technology |
High-throughput screening via FACS/MACS |
Scalable fermentation and purification processes |
|
Library Scale |
Up to 10⁸–10¹⁰ variants for discovery |
Single optimized construct for production |
|
Typical Outcome |
Improved binders, enzymes, or receptors |
Purified, active protein for downstream use |
|
Role in Workflow |
Early-stage discovery and optimization |
Final-stage expression and manufacturing |
How They Work Together
- E. coli Display Platform → Discover or evolve superior variants.
- E. coli Expression System → Express, purify, and characterize the optimized protein at scale.
Our Services & Capacities
Creative BioMart provides a comprehensive suite of services based on our validated E. coli display platform, covering every stage of the directed evolution pipeline—from library design to protein characterization. Our offerings include:
-
Selection of Carrier and Host Components
Identification of optimal coat proteins, signal peptides, and E. coli strains (e.g., BL21, TG1, or C41) for efficient surface expression.
-
Design and Implementation of Display Strategies
Using outer membrane proteins (OmpA, OmpC), flagellar fusions, or lipoprotein anchoring motifs to present peptides or complex proteins.
-
Validated Expression and High-Throughput Screening
Employing MACS or FACS for rapid enrichment of clones with improved binding, enzymatic activity, or stability.
-
Clone Amplification and Sequencing
Comprehensive genetic analysis and validation of selected mutants.
-
Recombinant Expression and Functional Analysis
Scalable production, purification, and biochemical characterization of evolved variants.
-
Cross-system Development
Extension of successful strategies to other Gram-negative bacteria such as Proteus mirabilis for specialized applications.
Service Workflow

Service Features
|
Parameter |
Specification |
|---|---|
|
Library Size |
Approximately 10⁸–10¹⁰ variants for robust diversity. |
|
Mutagenesis Approaches |
Both in vitro and in vivo mutagenesis options available. |
|
Display Strategies |
Outer membrane protein fusions (OmpA, LamB), flagellar display, and lipoprotein-based anchoring. |
|
Screening Technologies |
Automated FACS and MACS for precise and efficient selection. |
|
Host Systems |
Standard E. coli strains (BL21, TG1, C41) and alternative Gram-negative systems (e.g., Proteus mirabilis ). |
|
Target Applications |
Vaccine design, enzyme optimization, affinity maturation, receptor evolution, and ligand binding studies. |
Why Choose Creative BioMart
- Extensive Experience: Years of expertise in E. coli display and directed evolution, validated through numerous successful projects.
- Comprehensive Platform: From mutagenesis to characterization, we provide integrated, end-to-end protein engineering services.
- High Diversity Libraries: Up to 10¹⁰ variants, ensuring comprehensive coverage of sequence space.
- Advanced Screening Technologies: State-of-the-art MACS and FACS enable rapid identification of superior variants.
- Flexible Host and Display Options: Tailored anchoring motifs and host strains to suit target complexity.
- Cross-Species Adaptability: Expertise extending beyond E. coli to other Gram-negative systems for specialized applications.
Representative Case Studies
Case 1: An E. coli display method for characterization of peptide–sensor kinase interactions
Brink et al., 2023. Doi:10.1038/s41589-022-01207-z
Bacteria rely on two-component systems (TCSs) to detect peptides involved in host defense, quorum sensing, and inter-bacterial competition, yet the full range of TCS peptide-sensing remains poorly understood. Using an E. coli display method (SLAY-TCS), researchers characterized the effects of human antimicrobial peptides (AMPs) on the Salmonella Typhimurium PhoPQ TCS with unprecedented throughput. PhoPQ was found to sense AMPs with diverse sequences, structures, and functions. Combining thousands of AMP variants with machine learning revealed sub-domains and biophysical features linked to PhoPQ activation. SLAY-TCS reliably assessed positive and negative control peptides, including those forming disulfide bonds, demonstrating its utility for probing TCS specificity, mechanisms, and evolutionary dynamics.
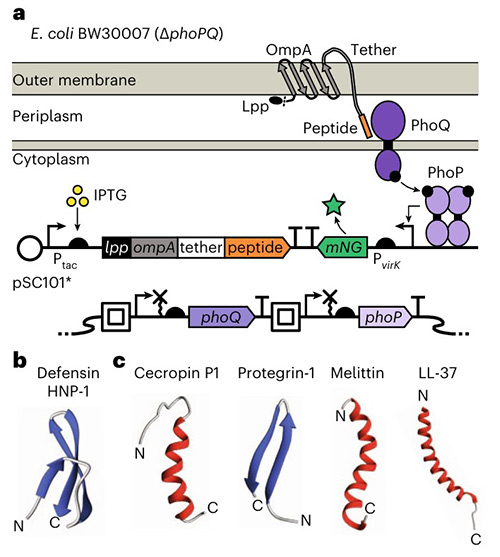
Figure 2. Surface-displayed AMPs activate PhoPQ in E. coli. a, Schematic of peptide surface display and genetic diagrams of peptide surface display and mNG fluorescent reporter plasmid. b, Structure of defensin HNP-1 (PDB ID, HNP-1: 3GNY), the only human AMP known not to activate PhoQ. (Brink et al., 2023)
Case 2: E. coli display enables high-affinity nanobody selection against cell surface antigens
Salema et al., 2016. doi:10.1080/19420862.2016.1216742
Therapeutic antibodies predominantly target cell surface proteins, but conventional phage display can be challenging for selecting binders on live cells due to the sticky nature of phages. Using an E. coli surface display system, researchers expressed VHHs (nanobodies) on bacterial surfaces and applied magnetic-activated cell sorting (MACS) and whole-cell selection (CellS) to isolate high-affinity clones against human EGFR. Screening of selected bacterial clones identified multiple VHHs binding distinct EGFR epitopes, including ligand-competitive clones. Flow cytometry and binding assays confirmed specificity, demonstrating that E. coli display enables efficient isolation and characterization of nanobodies targeting cell surface antigens, complementing traditional selection methods and expanding the antibody discovery toolkit.
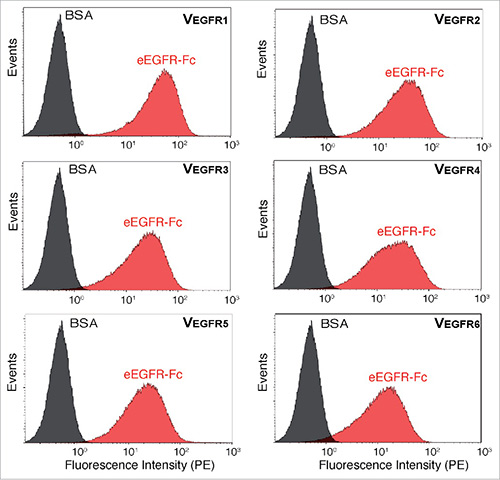
Figure 3. Binding of E. coli bacteria displaying selected VHH clones to eEGFR-Fc. Fluorescent flow cytometry analysis of induced E. coli EcM1 cells displaying the indicated Nb (VEGFR1-6) incubated with biotinylated eEGFR-Fc (red) or BSA (black) and secondary Streptavidin-PE. (Salema et al., 2016)
Client Reviews About Our E. coli Display Services
"Creative BioMart’s E. coli display system enabled us to evolve a key industrial enzyme with dramatically improved substrate selectivity. Their team designed a tailored surface-display strategy coupled with in vivo mutagenesis, allowing us to screen millions of variants efficiently. Within just eight weeks, they delivered a mutant with 12-fold higher catalytic efficiency and excellent thermal stability. The clear communication and comprehensive data package made the transition to our production line seamless."
— Director of Process Development | Global Industrial Biotechnology Company
"We collaborated with Creative BioMart to identify peptide ligands for a novel cytokine receptor target. Their E. coli display platform, combined with FACS-based high-throughput screening, allowed precise selection of binders with nanomolar affinity. The final peptides demonstrated exceptional receptor specificity in both binding and cell-based assays. Creative BioMart’s technical expertise and flexibility made them a trusted partner for our therapeutic discovery programs."
— Head of Protein Engineering | Mid-Size Biopharmaceutical Company
"Our group was struggling to express a GPCR in a functional form for binding studies. Creative BioMart applied their E. coli display system to direct the evolution of receptor variants with improved membrane expression and thermal stability. Their iterative screening approach yielded a construct with five stabilizing mutations, enabling us to finally perform crystallization trials. This achievement saved months of work and opened new structural biology opportunities for our research team."
— Principal Scientist, Structural Biology | Academic Research Institute
"We engaged Creative BioMart to screen an E. coli –displayed peptide library for antigenic epitopes relevant to our vaccine pipeline. Their team managed the entire process—from library design and MACS-based selection to clone sequencing and characterization. The top peptides identified showed high immunogenicity in our preclinical mouse model. Their scientific insight and consistent support throughout the project made a complex undertaking remarkably smooth."
— Director of Vaccine R&D | Global Pharmaceutical Company
FAQs About E. coli Display Platform
-
Q: What types of display strategies are available?
A: We provide multiple surface display strategies tailored to your target and project needs, including:- Fusion to outer membrane proteins (e.g., OmpA, LamB, or OmpC).
- Integration into cell surface structures such as flagella.
- Lipoprotein-based anchoring for stable and efficient presentation.
-
Q: What applications can benefit from E. coli display?
A: E. coli display is a versatile platform for directed evolution and protein engineering. It is ideal for:- Enzyme optimization for improved catalytic efficiency or substrate specificity.
- Affinity maturation of peptides, antibodies, and ligands.
- Vaccine candidate screening through antigen or epitope discovery.
- Membrane protein stabilization and receptor evolution.
- Ligand–receptor interaction mapping and functional analysis.
-
Q: How large are the libraries you can construct for E. coli display?
A: Our display libraries reach diversity levels up to 10⁸–10¹⁰ variants, ensuring broad sequence coverage and high discovery potential. Libraries can be created using random mutagenesis, site-directed mutagenesis, or in vivo mutagenesis. We can also incorporate customer-supplied sequences or custom-designed diversity to focus on specific binding or catalytic regions. -
Q: Which screening technologies are used for clone selection?
A: We employ both fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS) to rapidly enrich for the most promising variants. These high-throughput technologies enable precise control over selection stringency, allowing the isolation of clones with enhanced affinity, specificity, or activity in a fraction of the usual time. -
Q: Can your perform both in vitro and in vivo mutagenesis?
A: Yes. Our in vitro mutagenesis methods generate diverse variant libraries before transformation, while in vivo mutagenesis in E. coli continuously introduces mutations during growth, enabling ongoing diversification and adaptive evolution. This dual approach ensures both comprehensive coverage and practical efficiency for evolving complex proteins. -
Q: How does the E. coli display platform compare to other display systems like phage or yeast display?
A: E. coli display offers several advantages:- Direct link between genotype and phenotype for efficient selection.
- Fast growth and easy genetic manipulation for high-throughput applications.
- Compatibility with flow cytometry for quantitative screening.
- Broad host versatility—adaptable to other Gram-negative bacteria such as Proteus mirabilis.
-
Q: What deliverables will I receive at the end of an E. coli display project?
A: You will receive a comprehensive report and validated deliverables, including:- Library construction details and screening strategy.
- DNA sequences of selected clones.
- Binding, enzymatic, or stability assay results.
- Expression and purification data for evolved variants.
- Recommendations for further validation or scale-up.
Other Resources
Related Services
- Protein Engineering Services
- Directed Evolution
- Mutagenesis Services
- Protein Characterization
- Protein Expression and Purification Services
- Bacterial Expression Systems (E. coli / Bacillus)
- GPCR Screening Assays
- Transporter Screening Assays
- Large-Scale Protein Production Service
- High-Throughput Screening
- Enzyme Activity Assay
- Phage Display Platform
- Yeast Display Platform
- Special Cell-based Display Platform
- Cell-free Display Platform
Related Products
References:
- Brink KR, Hunt MG, Mu AM, et al. An E. coli display method for characterization of peptide–sensor kinase interactions. Nat Chem Biol. 2023;19(4):451-459. doi:10.1038/s41589-022-01207-z
- Lee SY, Choi JH, Xu Z. Microbial cell-surface display. Trends in Biotechnology. 2003;21(1):45-52. doi:10.1016/S0167-7799(02)00006-9
- Salema V, Mañas C, Cerdán L, et al. High affinity nanobodies against human epidermal growth factor receptor selected on cells by E. coli display. mAbs. 2016;8(7):1286-1301. doi:10.1080/19420862.2016.1216742
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

