Library Construction-Site Directed Mutagenesis
Library construction through site-directed mutagenesis is a cornerstone of modern protein engineering, enabling precise and efficient creation of mutant variants for structure–function analysis, enzyme optimization, and directed evolution. Creative BioMart provides comprehensive site-directed and saturation mutagenesis services tailored to library generation. With extensive experience in codon design, mutagenesis optimization, and sequence verification, our team ensures mutant libraries of high fidelity, minimal redundancy, and consistent diversity. Leveraging proprietary technologies and structural insights, we deliver ready-to-screen variant libraries that accelerate your protein discovery and functional characterization efforts.
Background: Site-Directed and Multisite-Directed Mutagenesis for Library Construction
Site-directed and multisite-directed mutagenesis are essential tools in protein engineering for generating targeted genetic variations at specific sites or defined regions within a gene. These methods enable precise exploration of structure–function relationships, allowing researchers to identify functionally important residues, enhance protein stability, or improve catalytic and binding properties.
This approach relies on structural and functional insights to guide mutation design. In certain cases, all possible amino acid substitutions are introduced at a particular site—a process known as saturation mutagenesis—to maximize structural diversity and uncover optimal residue configurations. Saturation mutagenesis typically employs synthetic oligonucleotides containing randomized codons flanked by wild-type sequences, allowing systematic codon-level exploration.
Several well-established methods are available for site-directed mutagenesis. Among them, the QuikChange method is widely used due to its efficiency and simplicity, eliminating the need for restriction enzyme digestion and ligation. For more complex applications, such as randomization of longer protein regions or simultaneous modification of multiple residues, advanced techniques like one-pot simple methodology for cassette randomization and recombination (OSCARR) or in vivo mutagenesis using E. coli vectors can be employed.
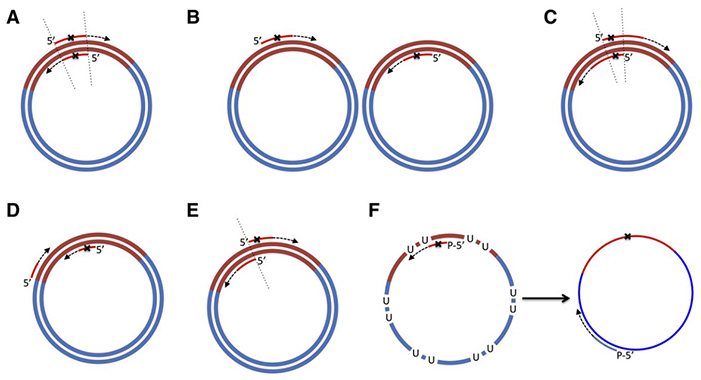
Figure 1. Primer design in original QuikChange method and QuikChange-derivatives. (Tee and Wong, 2013)
At Creative BioMart, we integrate rational mutagenesis design, custom oligonucleotide synthesis, and optimized cloning strategies to deliver high-quality variant libraries. Our platform supports both single-site and multisite designs with precise control over mutation frequency and codon diversity. The resulting mutant collections are ideal for high-throughput screening, functional characterization, and directed evolution studies, enabling researchers to efficiently explore and optimize protein performance.
Comprehensive Site-Directed Mutagenesis Services
Creative BioMart provides a suite of site-directed mutagenesis and library construction services designed to meet diverse protein engineering goals. Our capabilities include:
-
Site-Directed and Multisite-Directed Mutagenesis
Introduce single or multiple mutations with precision.
-
Saturation Mutagenesis
Achieve complete amino acid diversity at selected positions using degenerate oligonucleotides.
-
Custom Mutant Library Design
Tailored codon randomization schemes to balance diversity and screening throughput.
-
Sequence Validation
Sanger or NGS confirmation of construct integrity and variant distribution.
-
Low Background Wild-type Sequences
Ensuring minimal redundancy and truncated variants.
-
Flexible Delivery Formats
Plasmid DNA, expression-ready clones, or transformed cells.
Whether you aim to probe key active-site residues or generate an extensive functional variant library, Creative BioMart delivers precise, reproducible, and application-ready results.
Service Workflow
Our optimized workflow ensures accuracy, reproducibility, and fast turnaround for high-quality mutant library generation:
|
|
Step |
Description |
|
1 |
Project Consultation & Design |
Collaborative planning to define mutagenesis strategy, targeted residues, and desired diversity. |
|---|---|---|
|
2 |
Oligonucleotide Synthesis & Vector Preparation |
High-fidelity oligos with tailored degeneracy designed for optimal codon coverage and translation efficiency. |
|
3 |
Mutagenesis Reaction |
Performed using optimized site-directed or saturation mutagenesis protocols (e.g., QuikChange, OSCARR). |
|
4 |
Transformation & Library Construction |
Efficient cloning and transformation into E. coli or other host systems for variant expression. |
|
5 |
Screening & Sequence Validation |
Quality control through sequence analysis to confirm mutation frequency, diversity, and library representation. |
|
6 |
Delivery of Validated Libraries |
Final deliverables include plasmid DNA, expression constructs, and optional functional screening support. |
Multiple Mutagenesis Platforms
Creative BioMart employs multiple mutagenesis platforms to meet varied library construction needs:
- QuikChange-Based Mutagenesis: Ideal for 1–3 nucleotide substitutions without restriction enzyme or ligation steps.
- Improved QuikChange Variants: Used for enhanced efficiency and longer substitution stretches.
- OSCARR (One-Pot Simple Methodology for Cassette Randomization and Recombination): Enables larger region randomization or multiple contiguous site substitutions.
- In vivo Mutagenesis Systems: Vector-based strategies in E. coli hosts for continuous mutagenesis cycles.
Each library is designed for high mutation precision, minimal wild-type carryover, and optimized sequence diversity, ensuring superior quality for screening and functional evaluation.
Why Choose Our Site-Directed Mutagenesis Services
- Expertise in Rational and Random Mutagenesis: Decades of experience in designing codon randomization schemes guided by structural and functional insights.
- High-Fidelity Libraries: Proprietary cloning and screening protocols minimize wild-type and redundant sequences.
- Flexible Mutagenesis Platforms: From QuikChange to OSCARR and in vivo approaches, we adapt to your specific project goals.
- Comprehensive Validation: Every library is sequence-verified to ensure diversity and accuracy.
- Scalable Solutions: From small focused libraries to large-scale saturation mutagenesis projects.
- Fast Turnaround with Full Technical Support: Transparent communication, milestone updates, and expert consultation at each stage.
Case Studies and Real-World Applications
Case 1: Library construction and evaluation for site saturation mutagenesis
Sullivan et al., 2013. doi:10.1016/j.enzmictec.2013.02.012
A robust site-saturation mutagenesis method was developed to generate high-quality libraries, achieving an average of 27.4 ± 3.0 codons represented out of 32 in pools of 95 transformants. Verified across 11 independent OYE 2.6 gene libraries from Pichia stipitis , the protocol emphasized optimized PCR primer design and transformation efficiency as key factors for success. A quantitative analysis metric (Q-value) was also introduced to assess library degeneracy directly from fluorescence sequencing data. This early-stage quality evaluation accurately predicted final library diversity, enabling researchers to identify high-quality libraries early and efficiently prioritize those for detailed screening of functional protein variants.
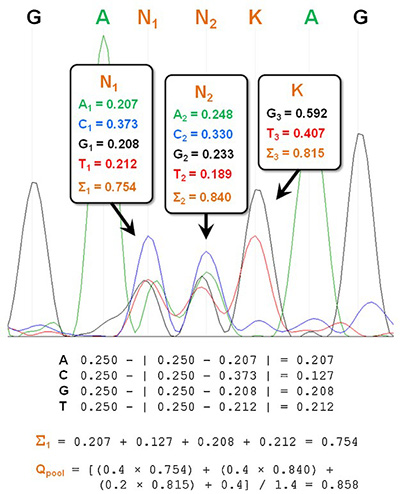
Figure 2. Method for quantitatively assessing library quality. Peak heights for each of the four fluorescence channels are measured at the randomized positions, then the fractional compositions of each base are calculated (A, green; C, blue; G, black, T, red). These values are subtracted from the theoretical composition (0.25 for N positions, 0.50 for K positions). Subtracting the absolute values of these deviations from the theoretical compositions, then summing these values yields ∑n values for each base position. The weighted average across the three bases of a codon yields and adding a scaling factor yields a Qpool value between 0 and 1. (Sullivan et al., 2013)
Case 2: Generation of an actagardine A variant library through saturation mutagenesis
Boakes et al., 2012. doi:10.1007/s00253-012-4041-0
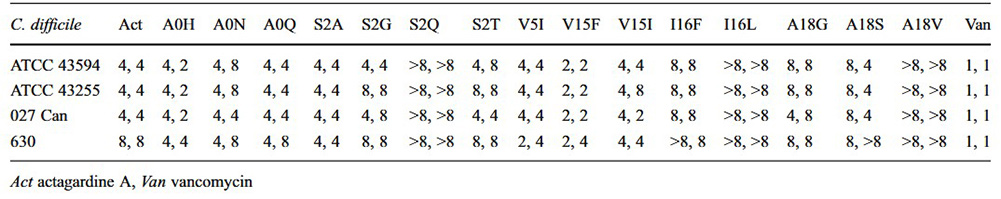
Actagardine A, a 19-amino-acid lantibiotic produced by Actinoplanes garbadinensis , exhibits potent antibacterial activity against major Gram-positive pathogens. To enhance its therapeutic potential, researchers developed a comprehensive site-directed mutagenesis system enabling targeted amino acid substitutions across the peptide, excluding residues involved in bridge formation. In total, 228 variants were engineered, with 44 successfully expressed in good yield. Notably, the V15F mutant displayed superior activity against multiple pathogens, including Clostridium difficile . This extensive variant library not only highlights the structural flexibility of actagardine A’s biosynthetic machinery but also provides a valuable foundation for designing improved lantibiotic analogs with enhanced antimicrobial properties.
Table 1 The antimicrobial activity of actagardine A substitution variants shown as minimum inhibitory concentrations (in micrograms per millilitre) against four strains of C. difficile. (Boakes et al., 2012)
Customer Testimonials on Site-Directed Mutagenesis
"We collaborated with Creative BioMart to generate a focused mutagenesis library targeting the ligand-binding pocket of a novel GPCR. Their team provided valuable input on codon randomization design and delivered a validated library with exceptional mutation coverage and minimal wild-type carryover. This allowed our internal screening team to identify multiple functionally distinct receptor variants within weeks. Their technical depth and responsiveness made the collaboration seamless from start to finish."
— Head of Molecular Pharmacology | Global Biopharmaceutical Company
"As part of an enzyme optimization program, we needed a reliable partner to construct saturation mutagenesis libraries for an industrial biocatalyst. Creative BioMart’s scientists not only guided us through oligonucleotide design but also provided detailed sequence validation data that confirmed >99% accuracy across the library. The resulting mutants helped us discover variants with improved thermal tolerance, significantly enhancing our process yield."
— Senior R&D Scientist | Industrial Biotechnology Company
"We engaged Creative BioMart to assist in constructing a multisite-directed mutagenesis library for an antibody fragment with improved binding kinetics. Their use of OSCARR methodology produced a diverse, high-quality library covering our targeted positions with excellent codon representation. Their fast turnaround and transparent communication made integration into our workflow effortless. The improved variant we identified has now advanced into our preclinical development pipeline."
— Director of Protein Engineering | Mid-Sized Biotech Firm
"Our structural biology lab needed a precise mutant library to study structure–function relationships in a membrane transporter. Creative BioMart’s team combined structural modeling insights with custom oligo synthesis to generate a clean, sequence-verified library. Their meticulous documentation and technical support helped our researchers rapidly identify functionally relevant mutations for crystallography trials. Their professionalism and scientific rigor rival that of top-tier core facilities."
— Principal Investigator | University Structural Biology Center
FAQs About Site-Directed Mutagenesis for Library Construction Services
-
Q: What types of mutagenesis strategies do you offer for library construction?
A: We provide a full range of mutagenesis solutions, including site-directed, multisite-directed, and saturation mutagenesis. Depending on your experimental goals, we use optimized protocols such as QuikChange-based mutagenesis for targeted substitutions or OSCARR for larger region randomization. Our team also offers in vivo mutagenesis systems using E. coli hosts for continuous mutational diversification. -
Q: How do you ensure the quality and diversity of the mutant library?
A: Every library is constructed with carefully designed oligonucleotides to achieve balanced codon coverage and minimal bias. After construction, we perform rigorous sequence validation—via Sanger or next-generation sequencing—to confirm diversity, reduce wild-type carryover, and eliminate truncated or redundant sequences. This ensures you receive a high-fidelity, screening-ready mutant library. -
Q: Can you customize mutagenesis projects for specific research goals, such as enzyme optimization or receptor profiling?
A: Absolutely. We tailor each project based on your target protein, functional objectives, and screening strategy. Whether you are performing active-site optimization, ligand-binding studies, or structure–function mapping, our team works closely with you to design targeted mutagenesis schemes that maximize the likelihood of identifying beneficial variants. -
Q: What makes your library construction service different from standard mutagenesis providers?
A: Unlike generic cloning services, our approach integrates rational mutagenesis design with structural and functional insights, ensuring purposeful rather than random diversity. Combined with our proprietary workflow for minimizing wild-type contamination and our high success rate in library generation, we deliver mutant collections that are both scientifically robust and application-ready. -
Q: What is the typical turnaround time for library construction projects?
A: Turnaround time depends on library complexity and sequence verification requirements. A focused site-directed library is typically delivered within 3–4 weeks, while large-scale saturation or multisite libraries may take 5–8 weeks. Every project includes progress updates and technical consultation throughout the process. -
Q: How are the libraries delivered, and in what formats?
A: We offer flexible delivery formats, including purified plasmid DNA, expression-ready clones, or transformed host cells (e.g., E. coli ). Libraries can also be provided with complete sequence verification reports and optional functional screening support upon request. -
Q: Do you assist with downstream applications such as screening or functional characterization?
A: Yes. While our primary focus is on library construction, we also offer optional downstream support—including assay development, protein expression, and functional screening services—to help accelerate your discovery pipeline. -
Q: How do I initiate a project or request a quote?
A: You can begin by contacting our technical support team with your gene or protein sequence, mutation targets, and desired library size. We’ll provide a free project consultation, discuss technical feasibility, and send a detailed quotation outlining the proposed workflow, timeline, and deliverables.
Other Resources
Related Services
- Protein Engineering Services
- Directed Evolution
- Mutagenesis Services
- Site-Saturation Mutagenesis
- Random Mutagenesis for Library Construction
- DNA Recombination
- Protein Sequence Analysis and Function Prediction
- Codon Optimization
Related Products
References:
- Boakes S, Ayala T, Herman M, Appleyard AN, Dawson MJ, Cortés J. Generation of an actagardine A variant library through saturation mutagenesis. Appl Microbiol Biotechnol . 2012;95(6):1509-1517. doi:10.1007/s00253-012-4041-0
- Sullivan B, Walton AZ, Stewart JD. Library construction and evaluation for site saturation mutagenesis. Enzyme and Microbial Technology . 2013;53(1):70-77. doi:10.1016/j.enzmictec.2013.02.012
- Tee KL, Wong TS. Polishing the craft of genetic diversity creation in directed evolution. Biotechnology Advances . 2013;31(8):1707-1721. doi:10.1016/j.biotechadv.2013.08.021
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

