Membrane-Based Yeast Two-Hybrid System
The Membrane-Based Yeast Two-Hybrid (MbY2H) system is an advanced protein interaction assay designed to overcome the limitations of conventional yeast two-hybrid (Y2H) techniques. Unlike traditional Y2H, which struggles with integral membrane proteins or self-activating nuclear proteins, MbY2H detects interactions directly at the membrane using a split-ubiquitin protein complementation approach. This method enables the investigation of full-length membrane proteins and membrane-associated proteins as baits, facilitating the discovery of novel interaction partners. Highly adaptable and compatible with high-throughput screening, MbY2H is an indispensable tool for comprehensive protein interaction studies in various organisms.
Introduction to Membrane-Based Yeast Two-Hybrid System
Understanding protein-protein interactions (PPIs) is fundamental to elucidating cellular pathways, signaling networks, and molecular mechanisms underlying physiological and pathological processes. Over the past decade, the classical yeast two-hybrid system and its modified variants have become invaluable for mapping protein interactions in a cellular context. This system relies on the reconstitution of a transcriptional activator when bait and prey proteins interact, allowing the activation of a reporter gene.
Despite its success, standard Y2H methods encounter significant challenges when studying certain classes of proteins. Integral membrane proteins, for example, are often retained within lipid bilayers, preventing proper nuclear localization required for classical Y2H readouts. Similarly, some nuclear proteins, including acidic transcription factors, may inherently activate the system even in the absence of genuine interactions, producing false-positive results. These limitations necessitated the development of alternative strategies that can accommodate challenging protein classes while maintaining high sensitivity and specificity.
The Membrane-Based Yeast Two-Hybrid system was developed to address these challenges. By detecting protein interactions directly at the membrane, MbY2H enables the study of full-length integral membrane proteins, membrane-associated proteins, and self-activating proteins in their native context. Leveraging the split-ubiquitin complementation principle, the system offers a robust platform for both targeted interaction studies and large-scale cDNA library screening to identify novel interaction partners.
Membrane-Based Yeast Two-Hybrid System: What We Offer
Creative BioMart provides a comprehensive Membrane-Based Yeast Two-Hybrid service tailored to the needs of academic and industrial researchers alike. Our offerings include:
-
Customized Experimental Design
Consultation to develop a strategy tailored to your proteins of interest, including selection of baits, preys, and reporter systems.
-
Cloning and Construct Preparation
Generation of bait and prey fusion constructs optimized for the split-ubiquitin system.
-
Interaction Screening
Single pairwise interaction validation or high-throughput screening against complex cDNA libraries.
-
Data Analysis and Interpretation
Comprehensive analysis of interaction results, including validation of positive hits and ranking of potential novel partners.
-
Flexible Service Formats
Options for academic research, pharmaceutical discovery, or biotech applications with scalable throughput.
By leveraging our expertise, clients can focus on biological insights while relying on a proven platform to uncover meaningful interactions.
Service Workflow

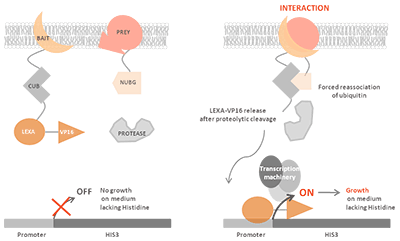
Principle of MbY2H
The MbY2H system exploits the split-ubiquitin complementation assay. Ubiquitin, a highly conserved small protein, can be divided into two stable fragments: the N-terminal half (Nub) and the C-terminal half (Cub). Neither fragment alone is sufficient to signal downstream events.
In this system:
- The bait protein is fused to Cub, which is linked to an artificial transcription factor.
- The prey protein is fused to Nub.
Upon interaction between bait and prey, the two ubiquitin halves reconstitute a pseudo-ubiquitin molecule. Cytosolic deubiquitinating enzymes recognize this molecule and cleave the transcription factor, which then translocates to the nucleus to activate reporter gene expression. This mechanism allows direct detection of interactions at the membrane, even for integral membrane proteins or proteins that would otherwise self-activate in classical Y2H assays.
-
MbY2H Advantages
- Compatible with full-length integral membrane proteins.
- Detects membrane-associated protein interactions.
- Accommodates self-activating transcription factors.
- Amenable to high-throughput cDNA library screening.
- Allows analysis in the native cellular membrane context, enhancing physiological relevance.
-
MbY2H Applications
- Mapping of membrane protein interactomes.
- Discovery of novel protein-protein interactions.
- Functional analysis of signal transduction pathways.
- Pharmaceutical target validation and drug discovery studies.
What Sets Us Apart
- Extensive Expertise: Decades of combined experience in protein interaction studies, particularly for challenging membrane and nuclear proteins.
- Customized Experimental Design: Tailored strategies to meet specific research objectives, ensuring optimal outcomes.
- Comprehensive Service: From cloning and yeast transformation to high-throughput screening and data analysis, all steps are handled in-house.
- High Sensitivity and Accuracy: Utilization of the split-ubiquitin system minimizes false positives and maximizes detection of true interactions.
- Scalable Solutions: Flexible service options suitable for single protein pairs or large-scale interaction network projects.
- Data-Driven Insights: Detailed analysis and reporting help clients interpret results and guide follow-up experiments.
Membrane-Based Yeast Two-Hybrid System: Case Studies
Case 1: Membrane-based yeast two-hybrid analysis of TSWV–Thrips interactions
Zhang et al., 2023 . doi:10.1111/1744-7917.13138
To uncover thrips proteins involved in Tomato spotted wilt orthotospovirus (TSWV) transmission, researchers applied a split-ubiquitin membrane-based yeast two-hybrid (MbY2H) system using the viral glycoprotein GN as bait. The screen identified 67 interacting proteins, with ApoD, Orai, and Obstructor-E-like isoform X2 (Obst) selected for deeper validation through MbY2H, β-galactosidase, and luciferase assays. Obst proved functionally significant: its silencing reduced viral acquisition by up to 40%, lowered virus titers, and cut transmission rates by more than half in both leaf disk and whole-plant assays. In contrast, ApoD and Orai knockdown showed no measurable effects. Findings highlight Obst as a key facilitator of TSWV acquisition and transmission in Frankliniella occidentalis.
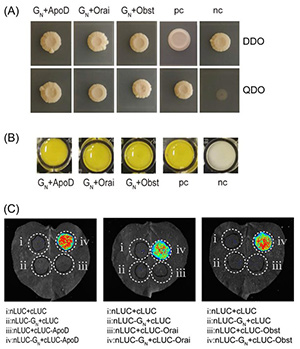
Figure 1. Validation of interactions between GN and ApoD, Orai, or Obst using MbY2H (A) and yeast β-galactosidase assays (B), and luciferase complementation assays (C). Interactions between pTSU2-APP and pNubG-Fe65 and between pTSU2-APP and pPR3-N were used as the positive control (pc) and the negative control (nc), respectively. DDO, double dropout medium (SD/-His/-Leu); QDO, quadruple dropout medium (SD/-His/-Leu/-Trp/-Ade). (Zhang et al., 2023)
Case 2: Membrane-based yeast two-hybrid mapping of MMV glycoprotein interactors
Barandoc-Alviar et al., 2022. doi:10.1101/2022.02.01.478665
To investigate how maize mosaic virus (MMV) invades and spreads within its insect vector Peregrinus maidis, researchers used a split-ubiquitin membrane-based yeast two-hybrid (MbY2H) system with the viral glycoprotein G as bait. The screen identified 125 interacting insect proteins, most linked to endocytosis, vesicle trafficking, protein turnover, nuclear transport, metabolism, and defense. Interaction networks suggested coordinated roles in G translation, folding, and trafficking. Cyclophilin A and apolipophorin III further co-immunoprecipitated and co-localized with G in insect cells. This work provides the first rhabdovirus spike-protein interactome in an insect vector, revealing host factors potentially essential for MMV infection and dissemination.
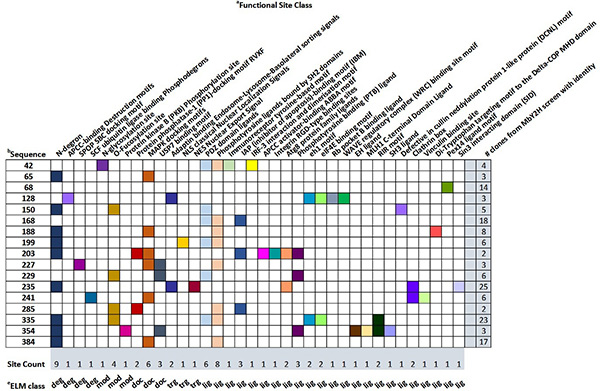
Figure 2. Prediction of functional sites (SLiMs) in nonannotated Peregrinus maidis proteins interacting with MMV-G in the MbY2H screen. (Barandoc-Alviar et al., 2022)
Membrane-Based Yeast Two-Hybrid System: Customer Feedback
“Creative BioMart’s MbY2H platform allowed us to uncover novel GPCR interactions that were previously inaccessible using traditional Y2H methods. Their team guided us through assay design, optimized bait constructs, and provided a detailed interaction map that has become foundational for our signaling pathway studies.”
— Director of Molecular Biology | Mid-Sized Biopharmaceutical Company
“We engaged Creative BioMart to screen a complex cDNA library against a set of membrane transporters involved in nutrient uptake. Their MbY2H workflow was highly efficient, and the validated hits they delivered directly informed our follow-up functional assays. The insights gained significantly accelerated our research timeline.”
— Senior Scientist, Plant Biology R&D | Agricultural Biotechnology Firm
“Our lab struggled with transcription factors that self-activate in classical Y2H systems. Creative BioMart’s MbY2H team implemented a modified approach that circumvented this issue entirely. The resulting interaction data were robust, reproducible, and instrumental in mapping transcriptional networks in human immune cells.”
— Principal Investigator | Academic Research Institute
“We needed a high-throughput platform to identify novel interactions for a family of ion channels relevant to cardiovascular disease. Creative BioMart not only executed the MbY2H screening flawlessly but also provided detailed data analysis and recommendations for follow-up validation. Their expertise gave our project team confidence in moving forward with target prioritization.”
— R&D Director, Cardiology Drug Discovery | Global Pharmaceutical Company
Membrane-Based Yeast Two-Hybrid System: Frequently Asked Questions (FAQs)
-
Q: What types of proteins can be analyzed using MbY2H?
A: The MbY2H system is specifically designed to handle challenging protein classes, including full-length integral membrane proteins, membrane-associated proteins, and self-activating transcription factors. Unlike classical yeast two-hybrid systems, MbY2H detects interactions directly at the membrane, making it ideal for proteins that are difficult to study in the nucleus or cause background activation. -
Q: Can MbY2H be used for high-throughput screening ?
A: Yes. The split-ubiquitin-based system is highly adaptable to high-throughput screening of complex cDNA libraries, enabling the discovery of novel interaction partners on a large scale. Our workflow supports both single-pair interaction studies and comprehensive library screening, with detailed data analysis provided for all hits. -
Q: How do you ensure the reliability of detected interactions?
A: Our platform uses a robust split-ubiquitin complementation approach, where only genuine bait-prey interactions lead to reporter activation. Additionally, we provide validation of positive hits and offer guidance on follow-up experiments to confirm biological relevance. This ensures minimal false positives and high confidence in the interaction data. -
Q: How customizable is the service for my project?
A: We offer fully tailored solutions. Our team will consult with you to design optimal bait and prey constructs, select the appropriate reporter system, and recommend modifications for self-activating proteins. Whether your project involves a single protein pair or a high-throughput library, we develop strategies to maximize the quality and relevance of your data. -
Q: What types of applications is MbY2H best suited for?
A: MbY2H is versatile and applicable to multiple research areas, including mapping membrane protein interactomes, discovering novel protein-protein interactions, studying transcription factor networks, and supporting pharmaceutical target validation or drug discovery. It is particularly valuable for projects where classical Y2H systems fail due to membrane retention or self-activation. -
Q: What is the typical workflow and turnaround time for a project?
A: Our standard workflow includes project consultation, construct preparation, yeast transformation or mating, interaction detection, high-throughput screening (if needed), and comprehensive data analysis. Turnaround times depend on project scale but are optimized for efficiency without compromising data quality. -
Q: Can you help interpret complex interaction data?
A: Absolutely. Beyond generating interaction results, our team provides detailed analysis, reporting, and interpretation of the data, including prioritization of potential interaction partners, functional insights, and recommendations for validation.
Other Resources
Related Services
References:
- Barandoc-Alviar K, Rotenberg D, Martin KM, Whitfield AE. Identification of interacting proteins of maize mosaic virus glycoprotein in its vector, Peregrinus maidis. Preprint posted online February 1, 2022. doi:10.1101/2022.02.01.478665
- Zheng X, Wan Y, Tao M, et al. Obstructor, a Frankliniella occidentalis protein, promotes transmission of tomato spotted wilt orthotospovirus. Insect Science. 2023;30(3):741-757. doi:10.1111/1744-7917.13138
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

