Nuclear Protein Extraction
Nuclear protein extraction is a critical technique for understanding the regulatory mechanisms that govern gene expression, signal transduction, protein–protein interactions, and post-translational modifications within the cell nucleus. At Creative BioMart, we provide high-purity nuclear protein preparations supported by optimized homogenization, advanced nuclear isolation strategies, and state-of-the-art analytical technologies. With decades of experience in subcellular proteomics, our team delivers reproducible, contamination-free nuclear extracts suitable for downstream applications such as tandem mass spectrometry, gel-based analyses, functional enzyme assays, epigenetic profiling, and transcription factor studies. Our comprehensive, customized workflows ensure precision, scalability, and data reliability for projects across academia, biotechnology, and pharmaceutical R&D.
Introduction to Nuclear Protein Extraction
Nuclear protein extraction is essential for studying transcription, chromatin regulation, DNA repair, and other nucleus-driven processes. Because nuclear proteins are often low in abundance and tightly associated with chromatin, obtaining high-purity extracts requires careful buffer design and precise handling. Standard workflows involve cold homogenization, filtration, centrifugation, and density-gradient separation to isolate intact nuclei, followed by optimized solubilization to preserve protein structure and post-translational modifications. Techniques such as Trizol-based extraction and tandem mass spectrometry then enable comprehensive profiling of nuclear proteins and their regulatory roles. With refined methods and extensive expertise, Creative BioMart delivers reliable, high-quality nuclear protein preparations for advanced proteomic studies.
Nuclear Protein Extraction: What We Offer
Creative BioMart provides comprehensive, customizable, and high-precision nuclear protein extraction solutions, including:
| Service |
Details |
|---|---|
|
Customized Homogenization Buffer Preparation |
Tailored buffer systems optimized for different tissues, species, developmental stages, and experimental conditions. |
|
Selection of Solubilization and Detergent Systems |
Strategies for effective membrane removal, chromatin dissociation, and protein solubilization without compromising integrity. |
|
Optimized Nuclear Isolation Procedures |
Full development and execution of isolation workflows featuring differential centrifugation, density gradients, and purification checkpoints. |
|
High-Purity Nuclear Protein Extraction |
Complete extraction services suitable for proteomics, epigenetic studies, transcription factor assays, and functional analyses. |
|
Protein Evaluation and Quality Assessment |
Verification through marker proteins, purity analysis, concentration determination, and compatibility testing for downstream applications. |
|
Mechanistic and Functional Study Support |
Integration with mass spectrometry, gel-based proteomics, post-translational modification analysis, and bioinformatic data interpretation. |
Our aim is to provide a one-stop solution that supports your research workflow from sample preparation to in-depth nuclear proteome characterization.
Service Workflow
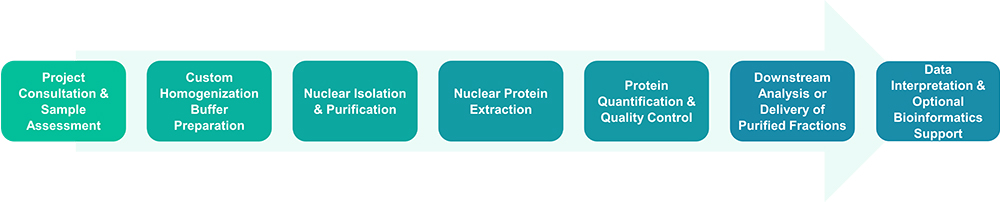
Service Features
-
Custom Buffer and Reagent Development
The choice of homogenization buffer significantly impacts the success of nuclear isolation. We develop customized formulations incorporating:
- Ionic or non-ionic detergents for selective membrane disruption
- Protease and phosphatase inhibitors to preserve protein stability
- Osmotic stabilizers to maintain nuclear morphology
- Reducing agents to protect sensitive post-translational modifications
Buffers are optimized for compatibility with subsequent extraction and MS workflows.
-
Advanced Nuclear Purification Strategies
We employ a full range of purification methods including:
- Differential centrifugation for rapid enrichment
- Discontinuous Percoll gradients for enhanced separation
- Continuous sucrose gradients for high-resolution purification
- Hybrid Percoll/sucrose systems for delicate or complex samples
- Continuous Percoll gradients for scalable purification
Each project receives a method uniquely optimized for tissue type, species, and research aims.
-
High-Purity Protein Extraction
Our extraction systems preserve:
- Protein–protein interaction complexes
- DNA- or chromatin-bound regulatory proteins
- Enzymatic activities
- Sensitive PTMs (phosphorylation, glycosylation, acetylation, S-nitrosylation)
Extraction conditions minimize protein degradation, preserve structural integrity, and maximize yield.
-
Proteomic and Functional Analysis
Available downstream services include:
- Tandem MS (LC–MS/MS)
- MALDI-TOF MS
- SDS-PAGE and 2-DE
- PTM mapping and quantification
- Nuclear enzyme activity assays
- Chromatin-associated protein studies
We provide comprehensive reports integrating proteomic profiles with functional or mechanistic insights.
What Sets Us Apart
- Decades of Experience in Nuclear and Subcellular Proteomics: Our long-standing expertise ensures efficient troubleshooting, optimized protocols, and reliable extraction of even the most challenging nuclear proteins.
- Tailor-Made Workflows for Any Species or Tissue Type: We specialize in custom buffer design and protocol optimization for mammalian tissues, plant tissues, microbial cells, and engineered cell lines.
- State-of-the-Art Analytical Technology: From high-resolution tandem MS to advanced gel-free proteomics, our platform delivers deep, accurate, and reproducible nuclear proteome data.
- High-Purity Preparation with Rigorous Quality Control: We validate nuclear fractions using multiple markers and provide detailed purity and contamination profiles to support research or publication needs.
- One-Stop End-to-End Service: Our integrated workflow—from sample processing to complex data interpretation—removes the need for multiple vendors and ensures consistency.
- Expert Support and Fast Turnaround: Our dedicated scientific team offers responsive communication, flexible customization, and timely delivery of results for projects of all scales.
Nuclear Protein Extraction: Case Studies
Case 1: Nuclear proteome analysis of salt-stressed Chlamydomonas reinhardtii
De Oliveira et al., 2022. doi:10.3390/phycology2030015
Microalgae, particularly Chlamydomonas reinhardtii, are promising feedstocks for biodiesel due to high neutral lipid productivity under abiotic stress. In this study, mixotrophic cultivation with 0.1 M NaCl rapidly increased intracellular neutral lipids without affecting growth, providing an efficient condition for bioenergy biomass production. Nuclear proteome analysis identified 323 proteins, with nearly 60% being nuclear, and 61 proteins were differentially regulated under salt stress. Key transcription factors and their regulatory elements were linked to lipid metabolism and kinase pathways, revealing nuclear mechanisms that drive lipid accumulation within the first 24 hours of salt exposure, offering insights for enhancing biofuel production.
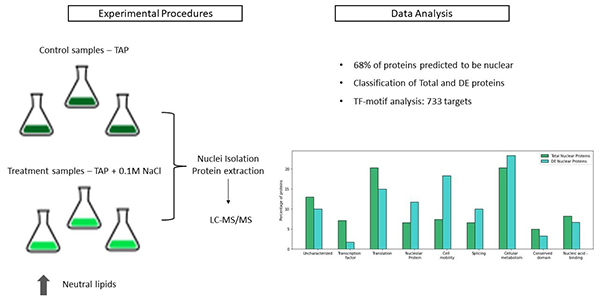
Figure 1. Characterization of the nuclear proteome of Chlamydomonas in response to salt stress. (De Oliveira et al., 2022)
Case 2: Nuclear protein extraction in amoebiasis research
Avila-Bonilla et al., 2025. doi:10.1016/j.exppara.2025.108965
Nuclear protein extraction played a central role in defining the nuclear proteome of Entamoeba histolytica, a major parasitic pathogen. Using mass spectrometry on purified nuclear fractions, researchers characterized the interactome of the nuclear polyadenylation factor EhCFIm25—an essential regulator of parasite proliferation and virulence. Extraction-based proteomics revealed unexpected enrichment of metabolic enzymes within the nucleus, suggesting moonlighting roles in gene expression and redox/energy coordination. The resulting RNA-binding and metabolic protein networks highlighted five key metabolic enzymes as potential regulators linking energy status to RNA metabolism. These findings provide new molecular targets and promising strategies for improved amoebiasis control.
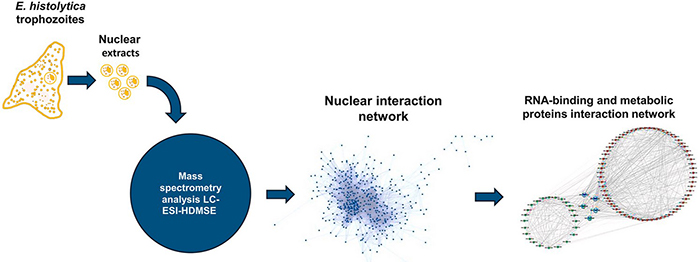
Figure 2. A network of RNA-binding and metabolic proteins evidenced in the Entamoeba histolytica nuclear proteome. (Avila-Bonilla et al., 2025)
Nuclear Protein Extraction: Customer Testimonials
“We engaged Creative BioMart to support a transcription-factor discovery program that required high-purity nuclear extracts from multiple human primary cell batches. Their team optimized a custom homogenization buffer for our unusually fragile cells and delivered consistently clean nuclear fractions with minimal cytoplasmic contamination. The downstream EMSA and ChIP-seq assays performed flawlessly on the first pass—something we rarely see. Their reliability significantly accelerated our internal validation timeline.”
— Director of Molecular Biology | Global Pharmaceutical Company
“Our project involved comparative nuclear proteome profiling across mouse, rat, and porcine tissues—historically a logistical headache due to the differences in nuclear stability among species. Creative BioMart standardized the extraction conditions, implemented tailored gradient centrifugation strategies, and delivered nuclear protein samples that were remarkably consistent in yield and purity. The LC-MS/MS data mapped thousands of nuclear proteins with confidence and uncovered several post-translational modifications we had previously missed. Their technical precision exceeded our expectations.”
— Principal Scientist, Proteomics | Biotechnology Research Institute
“We collaborated with Creative BioMart while characterizing the nuclear translocation behavior of a small-molecule inhibitor targeting chromatin-associated kinases. Their team generated high-integrity nuclear fractions suitable for both phosphorylation analysis and kinase activity profiling. Their meticulous documentation and repeatable workflow gave our regulatory group great peace of mind. The data they produced played a key role in our IND-enabling mechanistic package.”
— Senior Research Manager | Mid-Sized Therapeutics Company
“Our lab needed nuclear extracts from patient-derived tumor samples for histone modification and chromatin remodeling studies—a sensitive application where contamination can derail the entire experiment. Creative BioMart tailored the extraction protocol to our sample constraints and delivered nuclear proteins of exceptional purity with intact modification signatures. Their ability to preserve phosphorylation, acetylation, and methylation states made our downstream mass-spec mapping far more reliable.”
— Head of Genomics & Epigenetics | Academic Medical Center
Nuclear Protein Extraction Service: Frequently Asked Questions (FAQs)
-
Q: What types of samples can you process for nuclear protein extraction?
A: We handle a broad range of samples, including cultured cells, primary cells, and tissues from multiple species. Our team prepares custom homogenization buffers tailored to each sample type—particularly valuable for fragile cells or species with unique nuclear membrane properties—ensuring maximum yield and integrity of the nuclei. -
Q: How do you ensure high purity of nuclear fractions with minimal contamination?
A: Purity is achieved through carefully optimized isolation procedures, including customized solubilization strategies and advanced separation methods such as sucrose or Percoll gradient centrifugation. We validate nuclear purity using appropriate marker proteins to confirm nuclear enrichment and rule out cytoplasmic or organelle contaminants. -
Q: Can you preserve post-translational modifications during extraction?
A: Yes. Our workflows are specifically designed to maintain PTMs such as phosphorylation, acetylation, glycosylation, and S-nitrosylation. From inhibitor-supplemented buffers to controlled temperature processing, every step is optimized to keep modification profiles intact for downstream proteomic or mechanistic studies. -
Q: Do you support downstream analysis like mass spectrometry or functional assays?
A: Absolutely. Beyond extraction, we provide evaluation of nuclear proteins through gel-based analyses and advanced tandem mass spectrometry. For customers pursuing mechanism-of-action or signaling studies, we can also support follow-up functional assays or help design the most appropriate analytical approach. -
Q: How do you customize the extraction workflow for different projects?
A: Each project begins with a consultation to identify sample characteristics, required purity, intended assays, and key challenges. We then tailor homogenization buffers, select the appropriate detergents, choose the best density gradient method, and determine evaluation strategies so the final product aligns precisely with your research goals. -
Q: What quality controls are included with your nuclear protein extraction service?
A: We implement multiple QC checkpoints, including assessment of nuclear morphology, purity markers, protein concentration, and compatibility testing for downstream applications like ChIP-seq, EMSA, Western blotting, and MS-based proteomics. All QC data are documented thoroughly and delivered with your final report. -
Q: Can you scale up the extraction for large studies or multi-batch projects?
A: Yes. Whether you need a small pilot run or high-volume nuclear extraction across multiple tissue batches, our workflow is fully scalable. We maintain consistency through standardized protocols and controlled centrifugation parameters, allowing reproducible results across all sample sets.
Other Resources
Related Services
References:
- Avila-Bonilla RG, Velázquez Guzmán JA, Ramírez-Moreno E, Marchat LA. A network of RNA-binding and metabolic proteins evidenced in the Entamoeba histolytica nuclear proteome. Experimental Parasitology. 2025;274:108965. doi:10.1016/j.exppara.2025.108965
- De Oliveira Magalhães L, Nunes De Mello F, Vischi Winck F. Characterization of the nuclear proteome of Chlamydomonas in response to salt stress. Phycology. 2022;2(3):280-296. doi:10.3390/phycology2030015
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

