Phage Display Platform
Phage display is a cornerstone technology in directed evolution and protein engineering. By linking phenotype to genotype through bacteriophage display systems, it enables the selection of peptides, proteins, and antibodies with highly specific binding properties. Creative BioMart offers a robust, validated Phage Display Platform designed for the discovery and optimization of functional biomolecules. With our advanced in vitro and in vivo systems, we deliver comprehensive, high-throughput solutions from library construction to screening, clone amplification, and recombinant protein characterization.
Background: Understanding Phage Display
First described by George P. Smith in 1985, phage display revolutionized molecular biology by enabling the presentation of peptides or proteins on the surface of bacteriophages. The subsequent breakthrough in 1991, involving the selection of human antibodies using phage display, laid the foundation for therapeutic antibody discovery.
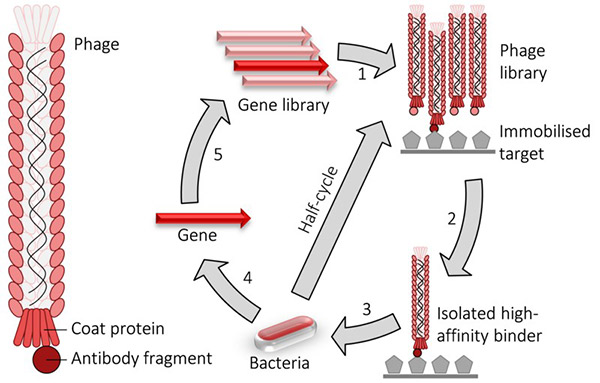
Figure 1. Phage display cycle.
Today, phage display libraries—containing up to billions of unique variants—are indispensable tools in the pharmaceutical industry. They facilitate the isolation of therapeutic antibody leads with exceptional specificity and affinity, particularly in the fields of oncology and immunology. A hallmark success of this technology is adalimumab, the world’s first fully human antibody derived via phage display, which achieved global recognition and annual sales surpassing $1 billion.
Phage display can be applied both in vitro and in vivo. In vitro systems typically involve the incorporation of genes encoding peptides or proteins into bacteriophage coat proteins (such as M13 pIII), enabling selection against a target molecule. In contrast, Phage- Assisted Continuous Evolution (PACE) —developed by David Liu and collaborators—permits continuous, in vivo evolution of proteins with complex functions, such as aminoacyl-tRNA synthetases.
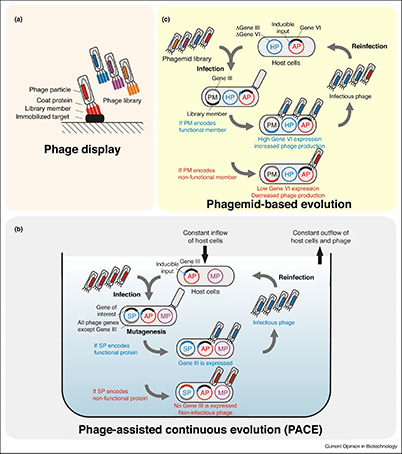
Figure 2. Phage-assisted directed evolution methods. (a) Affinity selection of library members by phage display. (b) Phage-assisted continuous evolution (PACE). (c) Phagemid-based evolution from combinatorial libraries in batch mode. (Brödel et al., 2018)
What Our Phage Display Planform Deliver
Creative BioMart has developed a validated, high-performance Phage Display Platform for directed evolution and molecular discovery. Leveraging years of technical expertise, we provide tailored solutions based on the target protein, desired function, and experimental goals.
Our comprehensive service package includes:
-
Selection of Optimal Coat Protein and Host System
We help you choose the most suitable phage and host combination to maximize display efficiency and binding fidelity.
-
Design and Implementation of Customized Phage Display Strategies
Every project begins with a tailored strategy to suit the target type and selection goal.
-
High-Throughput Screening for Rapid Enrichment
We employ advanced automation and assay systems for large-scale selection and validation of binding clones.
-
Amplification and Sequencing of Selected or Screened Clones
After successful selection, we isolate, amplify, and analyze the best-performing clones.
-
Recombinant Protein Expression, Purification, and Characterization
Our downstream services ensure that selected clones or evolved variants are fully characterized for functionality.
Service Workflow

Service Details
|
Parameter |
Specification |
|---|---|
|
Library Diversity |
Up to 10¹⁰ unique individual sequences for extensive coverage. |
|
Display Systems |
Featured M13 pIII peptide phage display platform for superior binding performance. |
|
Evolution Strategies |
Both in vitro and in vivo (PACE) protein evolution are available. |
|
Target Flexibility |
Multiple desired functions (binding, catalysis, stability) can be simultaneously optimized. |
|
Host Options |
Optimized E. coli and phage systems for high-efficiency amplification. |
|
Data Delivery |
Comprehensive reports including sequencing data, binding affinity analysis, and clone performance metrics. |
What Sets Us Apart
- Comprehensive One-Stop Service: From design to validation, we streamline every stage of phage display.
- Extensive Technical Expertise: Backed by years of successful directed evolution and antibody discovery projects.
- High Library Diversity: Up to 10¹⁰ variants ensuring exceptional discovery potential.
- Flexible Evolution Approaches: In vitro and in vivo systems to meet diverse research needs.
- High-Throughput Screening Capability: Rapid enrichment and validation of binders with advanced instrumentation.
- Proven Track Record: Trusted by global clients in pharmaceutical, biotech, and academic sectors.
Case Studies and Real-World Applications
Case 1: Optimizing a sarbecovirus nanobody via computational–phage display design
Ramos et al., 2025. doi:10.1073/pnas.2426438122
Combining computational design with phage display enables a powerful, targeted approach to broadening nanobody reactivity. While traditional phage display can screen up to 10¹⁰ variants, it still samples only a minute fraction of possible antibody–antigen interfaces. By integrating residue-specific pharmacophore modeling with directed phage display, researchers can focus mutational diversity toward the most promising binding residues. Applying this interdisciplinary method, a camelid nanobody (VHH-72) was redesigned to cross-react with multiple SARS-CoV strains, yielding a triple mutant with up to 20-fold enhanced affinity. This strategy showcases how computational guidance can significantly expand the functional landscape of single-domain antibodies.
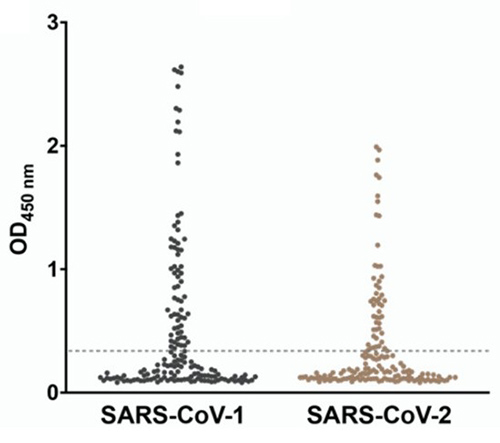
Figure 3. Binding reactivity of diverse VHH-72 phage clones toward the RBD from SARS-CoV-1 and SARS-CoV-2 characterized by the monoclonal phage ELISA. The dotted line represents the cutoff for clones displaying functionality. (Ramos et al., 2025)
Case 2: Phage-assisted continuous evolution of proteases with altered substrate specificity
Packer et al., 2017. doi:10.1038/s41467-017-01055-9
Using phage-assisted continuous evolution (PACE), researchers reprogrammed the TEV protease, which normally cleaves the ENLYFQS sequence, to instead recognize and cleave a distinct HPLVGHM motif within human IL-23. After approximately 2,500 generations, the evolved protease acquired 20 adaptive mutations and successfully inhibited IL-23–mediated immune signaling in murine splenocytes. Detailed specificity profiling revealed broadened and shifted substrate preferences, illuminating the molecular basis of altered recognition. This study demonstrates the power of PACE to re-engineer protease specificity within practical time frames, paving the way for designing custom proteases for therapeutic and biotechnological applications.
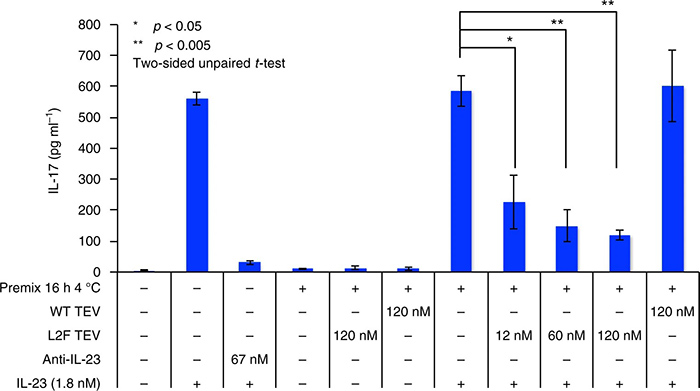
Figure 4. Protease-mediated attenuation of IL-17 secretion in mouse splenocytes. (Packer et al., 2017)
Customer Feedback on Phage Display Platform
“Creative BioMart’s phage display team helped us identify a panel of high-affinity antibodies against a novel immune checkpoint target within weeks. Their optimized M13 pIII system and automated screening pipeline significantly shortened our discovery timeline. The top clones they delivered exhibited sub-nanomolar affinity and exceptional selectivity, validated independently by our internal SPR assays. Their technical communication was prompt, transparent, and highly collaborative throughout the project.”
— Director of Biotherapeutics Research | Mid-Size Pharmaceutical Company
“We collaborated with Creative BioMart to evolve aminoacyl-tRNA synthetase variants with improved substrate tolerance using their in vivo Phage-Assisted Continuous Evolution (PACE) platform. Their scientists designed a clever mutagenesis and selection workflow that yielded functional variants with nearly 20-fold enhanced catalytic efficiency compared to our wild-type enzyme. The final deliverables were well-documented and included detailed kinetic characterization, saving our team months of internal optimization.”
— Head of Synthetic Biology Division | Biotechnology Startup
“Our R&D team needed to identify peptide ligands capable of selectively binding to tumor-associated receptors for a nanoparticle drug delivery program. Creative BioMart constructed a high-diversity peptide phage display library (10¹⁰ complexity) and conducted several stringent panning rounds. The shortlisted peptides demonstrated remarkable receptor specificity and stability in serum assays. Their scientists not only provided the sequences but also supported follow-up conjugation studies, which was invaluable to our project success.”
— Senior Scientist, Drug Delivery Research | Global Biopharmaceutical Company
“When a new viral strain emerged, speed was critical. Creative BioMart’s antibody phage display platform enabled us to isolate potent neutralizing antibody fragments against the viral surface antigen in record time. Their team customized the selection parameters to mimic physiological binding conditions, ensuring clinical relevance. The scFv clones showed strong neutralization in our pseudovirus assay and formed the foundation of our rapid diagnostic and therapeutic development efforts.”
— Principal Investigator, Infectious Disease Research | National Research Institute
FAQs About Phage Display Platform
-
Q: What types of phage display systems do you use?
A: We offer both M13 filamentous phage and T7 lytic phage display systems. Depending on your target molecule and project goal, we select the optimal coat protein (such as pIII or pVIII) and display format. Our featured M13 pIII peptide phage display platform is ideal for antibody and peptide discovery, while T7-based systems are preferred for higher expression or enzymatic targets. -
Q: Can you perform both in vitro and in vivo protein evolution?
A: Yes. We provide both in vitro selection (traditional panning for binders) and in vivo continuous evolution (PACE) for more complex functional optimization. The latter allows rapid, iterative mutation and selection within living cells, ideal for evolving enzymes, tRNA synthetases, or regulatory proteins with improved or novel functions. -
Q: How large are your phage display libraries, and can you use customer-supplied libraries?
A: Our in-house libraries reach a diversity of up to 10¹⁰ unique variants, covering a broad sequence and structural space. We can also work with customer-supplied or custom-built libraries, including antibody fragments (scFv, Fab, VHH), random peptides, or cDNA/ORF-based collections. Each library undergoes quality verification before use to ensure representation and stability. -
Q: What expression systems are available for recombinant protein production?
A: We support a full range of expression systems for downstream validation and production, including:- E. coli (high-yield bacterial expression)
- Pichia pastoris (yeast system for proper folding)
- HEK293 or CHO cells (for mammalian expression and post-translational modification)
- Baculovirus-insect cell systems
-
Q: How do you ensure the specificity and affinity of selected clones?
A: We combine iterative panning, high-throughput screening, and biophysical validation to guarantee the quality of selected clones. Each candidate undergoes binding affinity testing using ELISA, SPR, or BLI, as well as cross-reactivity analysis to ensure high specificity. For affinity maturation projects, we employ targeted mutagenesis and re-screening to further enhance performance. -
Q: What data and deliverables will I receive at the end of a project?
A: Clients receive a comprehensive project report including:- Library design and screening workflow
- Clone sequences and alignment data
- Binding affinity and specificity results
- Recombinant expression and purification records
- Functional assay results (if applicable)
-
Q: How long does a typical phage display project take?
A: Project timelines depend on complexity and target type. Standard antibody or peptide discovery projects typically require 6–10 weeks, while enzyme evolution or PACE-based studies may take 8–12 weeks. We maintain transparent communication throughout the process and can expedite timelines when necessary.
Other Resources
Related Services
- Protein Engineering Services
- Directed Evolution
- Protein Expression and Purification Services
- Bacterial Expression Systems (E. coli / Bacillus)
- Baculovirus-Insect Cell Expression
- Yeast Expression Systems
- Mammalian Expression Systems
- Epitope Mapping
- High-Throughput Screening
- Escherichia coli Display Platform
- Yeast Display Platform
- Special Cell-based Display Platform
- Cell-free Display Platform
Related Products
References:
- Brödel AK, Isalan M, Jaramillo A. Engineering of biomolecules by bacteriophage directed evolution. Current Opinion in Biotechnology. 2018;51:32-38. doi:10.1016/j.copbio.2017.11.004
- Packer MS, Rees HA, Liu DR. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat Commun. 2017;8(1):956. doi:10.1038/s41467-017-01055-9
- Ramos KE, Nandigrami P, Najafian A, Lai JR, Fiser A. Optimization of a sarbecovirus llama nanobody–antigen binding interface via a combined computational and phage display protein engineering approach. Proc Natl Acad Sci USA. 2025;122(28):e2426438122. doi:10.1073/pnas.2426438122
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

